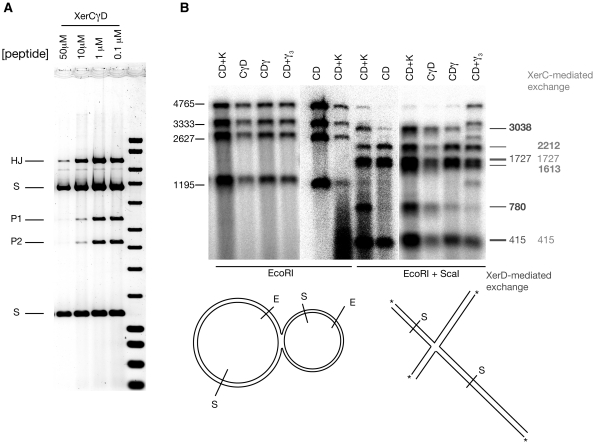
Figure 5.
(A) Recombination in the presence of peptide WRWYCR traps HJs. Recombination was carried out in the presence of the indicated concentration of peptide and subsequently cut with EcoRI so that HJs migrate slowly. (B) Denaturing alkali gels allow determination of exchanged strands in isolated HJs. Isolated HJs were 5′-end labelled at each EcoRI cut site. Subsequently, some of the DNA was then further digested with ScaI, and samples were then denatured and electrophoresed. The relative positions of each site are shown diagrammatically below the gel. Sizes of the four strands resulting from EcoRI digestion are shown alongside (left). The expected sizes of top strand exchange (XerC-mediated) and bottom strand exchange (XerD-mediated) are shown on the right. Two strand sizes (3038 and 780) are specific for XerD mediated exchange (shown in bold), while XerC mediated exchange produces two different diagnostic product sizes (2212 and 1613, also in bold). The other strand sizes (415 and 1727) are common to both events. Note that there is always a background of XerC-mediated exchange, which can be estimated from the CD alone lane. However, upon stimulation by γ (in any form) the level of XerD-mediated exchange is greatly increased. Note that there was partial digestion by ScaI of the XerCD + γ3 reaction (far right lane) so that the four bands seen with EcoRI digestion are still present.