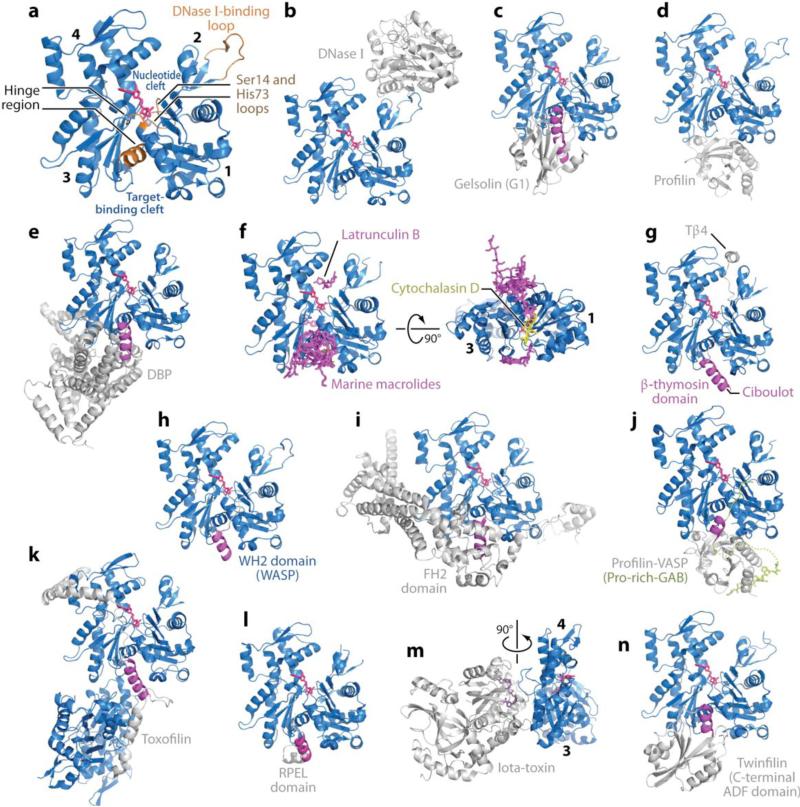
Figure 1.
Structures of actin and actin complexes. The structures of actin complexes are shown to scale and in chronological order of publication. (a) Classical view of the structure of the actin monomer. The structure shown was derived from the complex with DNase I, with completion of the C terminus from the complex of actin with profilin. Highlighted in orange are the Ser14 and methylated His73 loops, the DNase I-binding loop, and the hinge between domains, consisting of helix Gln137-Ser145 and the loop centered at residue Lys336. Subdomains 1--4 are labeled. (They are also labeled in panels f and m, which show rotated views of the structure.) Together, subdomains 1 and 2 form the outer (or small) domain, whereas subdomains 3 and 4 constitute the inner (or large) domain. Two large clefts are formed between these domains: the nucleotide- and target-binding clefts. Most actin-binding proteins (ABPs) and small molecules bind in the target-binding cleft, and the interaction frequently involves an α-helix (magenta). (b) DNase I (1ATN). (c) Gelsolin segment 1 (G1) (1EQY). See also Supplemental Figure 1 for structures of actin with Gelsolin fragments G1--G3 and G4--G6. (d) β-actin-profilin (2BTF). (e) Vitamin D-binding protein ( DBP ) (1KXP). (f) Two perpendicular views of a superimposition of structures of actin complexes with small molecules, including marine toxins (1QZ5, 1QZ5, 1S22, 1YXQ, 2ASM, 2ASO, 2ASP, 2FXU, 2Q0R, 2Q0U, 2VYP), Latrunculin B [DOUG: Add “A/B’ – missing.] (2Q0U) and cytochalasin D (3EKS). The marine toxins (magenta) bind at the ends of the target-binding cleft, whereas cytochalasin D binds in the middle, and Latrunculin (both A and B) binds in the nucleotide cleft. All these molecules compromise actin polymerization. (g) β-thymosin domain. A complete structure of this complex is not available, but combined, the structures of the N-terminal portion of a β-thymosin domain from Drosophila ciboulot (1SQK) and the C-terminal end of b-thymosin peptide (Tβ4) (1T44) provide a model of this complex. (h) WASP homology domain 2 (WH2) domain of WASP (2A3Z). The WH2 domain, present in many cytoskeletal proteins in the form of tandem repeats, is related to the β-thymosin domain but lacks the C-terminal pointed end capping helix. (i) Formin homology 2 (FH2) domain (1Y64). See also Supplemental Figure 2 for a more detailed representation of this structure. (j) Ternary complex with profilin and the Pro-rich G-actin-binding (Pro-rich-GAB) domains of VASP (2PBD). The GAB domain is related to the WH2 domain but presents a shorter N-terminal helix and adopts a slightly different orientation when bound to actin, possibly because it is designed to co-bind with profilin. (k) Toxofilin from Toxoplasma gondii bound to an antiparallel actin dimer (2Q97). (l) RPEL (RPxxxEL-containing motif)[AU: Does this need to be spelled out for the general reader?] domain from the serum response factor coactivator MAL (2V52). (m) Arginine ADP-ribosylation iota-toxin from Clostridium perfringens (3BUZ). View rotated 90° relative to the other complexes. (n) C-terminal ADF/cofilin domain of twinfilin (3DAW). See Supplemental Table 1 for a complete list of references.