Abstract
Gender differences in brain development and in the prevalence of neuropsychiatric disorders such as depression have been reported. Gender differences in human brain might be related to patterns of gene expression. Microarray technology is one useful method for investigation of gene expression in brain. We investigated gene expression, cell types, and regional expression patterns of differentially expressed sex chromosome genes in brain. We profiled gene expression in male and female dorsolateral prefrontal cortex, anterior cingulate cortex, and cerebellum using the Affymetrix oligonucleotide microarray platform. Differentially expressed genes between males and females on the Y chromosome (DBY, SMCY, UTY, RPS4Y, and USP9Y) and X chromosome (XIST) were confirmed using real-time PCR measurements. In situ hybridization confirmed the differential expression of gender-specific genes and neuronal expression of XIST, RPS4Y, SMCY, and UTY in three brain regions examined. The XIST gene, which silences gene expression on regions of the X chromosome, is expressed in a subset of neurons. Since a subset of neurons express gender-specific genes, neural subpopulations may exhibit a subtle sexual dimorphism at the level of differences in gene regulation and function. The distinctive pattern of neuronal expression of XIST, RPS4Y, SMCY, and UTY and other sex chromosome genes in neuronal subpopulations may possibly contribute to gender differences in prevalence noted for some neuropsychiatric disorders. Studies of the protein expression of these sex- chromosome-linked genes in brain tissue are required to address the functional consequences of the observed gene expression differences.
Keywords: RPS4Y, UTY, SMCY, XIST, DBY, USP9Y, VCX, VCY
INTRODUCTION
Gender differences have been noted in the prevalence of neuropsychiatric disorders such as depression, attention deficit hyperactivity, and drug abuse (American Psychiatric Association, 1994). Differences between normal development and aging of male and female brain have been noted in neuroimaging studies (Escalona et al, 1991; Giedd et al, 1996; Gur et al, 1991; Xu et al, 2000). Subtle gender differences in correlations between the size of brain structures and intelligence have been reported (Andreasen et al, 1993). Differences in neuron size and neuropil volume in the cortex of male and female controls have been reported in post-mortem stereological investigations (Pakkenberg et al, 2003; Rabinowicz et al, 2002).
There is evidence that nonsteroidal influences promote gender differences on a cellular basis in brain tissue. The number of dopaminergic midbrain neurons during development in mice can be independent of the effects of testosterone or estrogen, and related to expression of sex chromosome genes (Carruth et al, 2002). The expression of two Y chromosome genes in human brain, SRY (sex determining region Y) and ZFY, suggests that Y chromosome gene expression in adult brain might play some role in gender differences (Mayer et al, 1998). ZFY and SRY were found in frontal cortex and hypothalamus of males, but not in the temporal cortex of males, and both were absent in all brain regions of females (Mayer et al, 1998). The mRNA expressions of Y-linked genes (Usp9y, Ube1y, Smcy, Eif2s3y, Uty, and Dby) are found exclusively in male mouse brain and the X-chromosome gene, XIST, is expressed only in female mouse brain (Eriksson et al, 1999; Xu et al, 2002). The X-linked homologues (Usp9x, Ube1x, Smcx, Eif2s3x, Utx, and Dbx) were also expressed in females at significantly higher levels than males (Xu et al, 2002).
As part of an on-going study of neuropsychiatric disorders, we have chosen to focus our investigation of gender differences in gene expression to known biological differences due to sex chromosome linked genes. Upon detection of X- and Y-linked genes in human brain by microarray technology, we next asked if these genes are differentially or ubiquitously expressed in human brain across cell types, neuron vs glia, using in situ hybridization histochemistry. We also wished to investigate whether expression of X- and Y-linked genes was heterogeneous between brain regions and examined the question of regional differences in sex chromosome gene expression. On a technical standpoint, we also examine the sequence homologies between X- and Y-linked genes that could give rise to ‘cross-reactivity’. The technical issues surrounding probe set hybridization in male and female samples might also determine the ultimate success of the microarray approach to detection of gene expression differences. Finally with microarray studies, it is necessary to validate the findings with other techniques, and we demonstrate confirmation using quantitative real-time PCR and in situ hybridization.
METHODS
Demographics and Selection of Brain Samples
A total of 20 brains were obtained at autopsy from the Orange County Coroner’s Office through the Brain Donor Program at the University of California, Irvine, Department of Psychiatry (for a description of handling and processing the brains, see Jones et al, 1992). The cadavers were placed in a cold room prior to autopsy, decreasing the postmortem interval (PMI) at room temperature. For the microarray and TaqMan assay, 10 subjects (five males and five females) were group matched on the basis of age, brain pH, and interval between death and freezing the brain. The cause of death, the incidence of smoking, and drug abuse were similar between groups (Table 1, upper half). An additional 10 brains (six males and four females) were used for an independent confirmation by SYBR Green real-time PCR of gene expression differences (Table 1, lower half).
Table 1.
Demographic Characteristics of Group Matched Male and Female Subjects used in Microarray and TaqMan PCR Study and the Additional Subjects for SybrGreen PCR study
| Gender | Age | PMI | Frozen | Substance abuse | Cause of death | Tobacco use | pH |
|---|---|---|---|---|---|---|---|
| Microarray and TaqMan assay | |||||||
| F | 75 | 14 | 8/23/1991 | No | Cardiac | No | 6.13 |
| F | 53 | 17 | 10/30/1996 | No | Asthma | Hx | 6.98 |
| F | 82 | 28 | 3/14/2000 | No | Cardiac | Yes | 6.71 |
| F | 75 | 21 | 11/29/1999 | No | Cardiac | No | 6.4 |
| F | 69 | 21 | 10/17/2000 | No | Cancer | No | 6.12 |
| M | 71 | 9.8 | 3/10/1998 | No | Cardiac | No | 6.86 |
| M | 57 | 14 | 11/6/1991 | No | Cardiac | No | 6.94 |
| M | 82 | 30.75 | 8/1/2000 | No | Cancer | Yes | 6.49 |
| M | 72 | 24.5 | 5/19/1998 | No | Cardiac | No | 6.84 |
| M | 68 | 23 | 6/26/2000 | Hx | Cardiac | Yes | 6.43 |
| SybrGreen PCR assay | |||||||
| F | 47 | 41.3 | 4/23/2001 | No | Cancer | No | 6.73 |
| F | 64 | 27 | 5/19/2001 | No | Cancer | Hx | 6.15 |
| F | 68 | 25 | 6/13/2001 | No | Cancer | No | 6.82 |
| F | 73 | 24 | 7/5/2001 | No | Lung CA | Yes | 6.54 |
| M | 45 | 21 | 1/8/1998 | No | Cardiac | Yes | 6.86 |
| M | 52 | 23 | 11/29/1988 | Yes | Cancer | Yes | 6.06 |
| M | 50 | 16.5 | 2/5/2000 | No | Ulcer | Yes | 6.27 |
| M | 70 | 28 | 3/7/2000 | No | ASHD | No | 7.03 |
| M | 58 | 15.3 | 1/27/2001 | Yes | Cancer | Hx | 6.68 |
| M | 44 | 23 | 4/3/2001 | No | Cardiac | Yes | 6.87 |
PMI includes time from death to brain removal, cooling to 4ºC, and freezing of brain.
Hx, history of use/abuse.
Total RNA Extraction
Three regions from the left side of each brain were chosen for Trizol® (Invitrogen) extraction of total RNA: the anterior cingulate cortex (area 24), the dorsolateral prefrontal cortex (DLPFC) (area 9 plus 46), and the cortex of the cerebellar hemisphere. The extracted samples were retained for use when two ribosomal RNA bands (28S and 18S) were evident on ethidium bromide-stained agarose gels and the 28S band was more prominent than the 18S band. The total RNA sample for the anterior cingulate cortex, DLPFC, and cerebellum was divided into three aliquots. One aliquot of each total RNA samples was sent to three different laboratories for further handling and assay. As an additional check for RNA integrity after extraction of the cerebellar cortex, total RNA was analyzed by Agilent 2100 Bioanalyzer for 28S and 18S quantitation according to the manufacturer’s protocol. Total RNA samples were further purified by passing over silica-based mini-spin columns (Qiagen RNeasy Mini Kit, Chatsworth, CA) and eluting in 30–50 μl of DEPC water. Human testis total RNA was obtained from a commercial source for use in RT-PCR experiments (Ambion, TX) as a positive control to confirm the Y chromosome expression.
Oligonucleotide Microarrays
The labeling of total RNA, chip hybridization, and scanning of oligonucleotide microarrays were carried out at three independent sites (University of California, Irvine; University of California, Davis; University of Michigan, Ann Arbor) following a standard Affymetrix protocol (see Supplementary Method 1). Thus, there were experimental triplicates performed for each biological sample. There were 30 biological samples distributed to each of three sites creating a total of 90 microarray chips. The Qiagen column purified total RNA (10 μg) from each brain region was converted to biotinylated cRNA hybridization probe, following the manufacturer’s protocol (Affymetrix, Santa Clara, CA) at each of three sites.
Condensation of Probe Level Data from Affymetrix Oligonucleotide Arrays
The scanned image of the Affymetrix HGU95a array contains 12 625 probe sets with 16 perfect match and 16 mismatch probes contained in a single probe set. This raw probe level data was analyzed with dCHIP software (Li and Wong, 2001), which incorporates a statistical model for oligonucleotide expression array data at the probe level. The dCHIP algorithm can detect probe pairs that are not informative such as cases where the mismatch feature gives a stronger signal than the perfect match, or where possible artifacts due to cross-hybridization of a probe occurs independent of total RNA concentrations across samples. We used dCHIP for gender comparisons by t-test. We computed the signal for each site’s chips (n = 30 chips) with a dCHIP analysis using the perfect match minus mismatch algorithm. The gene expression differences for gender were then evaluated with a t-test. Since each of three sites ran all 30 samples, there were 90 microarray chips. Each set of 30 chips formed one experimental replicate, the set of 90 microarray chips represents three experimental replicates. The strategy we adopted was to analyze each experimental replicate by an independent dCHIP analysis. There were three independent dCHIP analyses of gene expression, and only genes that passed the three experimental replications are reported. The entire set of 30 chips represents a global analysis of gene expression across three brain regions, that is, lumping all three brain regions into one analysis.
A secondary analysis of gender differences in brain regions was conducted with separate dCHIP analyses for regional gene expression differences by gender. This resulted in nine dCHIP analyses, which were formed from three regions assayed in experimental triplicates.
To evaluate a fold-change and t-test criteria for significant gene expression differences, technical replicates of chips were used in a pilot study. Three DLPFC total RNA samples were labeled according to the standard Affymetrix protocol. The three fragmented cRNA hybridization cocktails were split and hybridized to two separate chips forming technical replicates. The analysis of these six chips in a pilot study was used to establish a baseline false positive threshold. When using the empirical criteria of both a ± 1.5-fold change and t-test of p < 0.05 on identical cRNA samples (technical duplicates), there was an average false positive rate of three genes in 12 625 probe sets that were significantly changed with these criteria.
The most significant genes following dCHIP analysis including either the top and bottom 5% of p-value ranking and fold changes were then classified using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database assignments (Ashburner et al, 2000; Kanehisa and Goto, 2000). The probability that classifications were over-represented in the significant gene lists was determined using GO (biological process, molecular function, and cellular component) and KEGG pathways. The website http://apps1.niaid.nih.gov/David/upload.asp provides annotation to convert Affymetrix probe sets to Locus Link identifiers and GO and KEGG pathways (Dennis et al, 2003).
The Affymetrix HGU95a dataset that was derived from comparisons of multiple tissue types (Su et al, 2002) was consulted for comparison of testicular and ovarian expression of selected genes (http://expression.gnf.org/cgi-bin/index.cgi) using the online web-based interface ‘Gene Expression Atlas’.
Quantitative PCR Assay with TaqMan Probe
A reverse transcription was run on total RNA extracted from the 10 samples from DLPFC in cohort I (five males and five females). Quantitative PCR reactions were carried out on the resulting cDNA using an Applied Biosystems 7700 and 7000 sequence detection systems (ABI, Foster City) according to the manufacture’s protocol in a 25 μl reaction. Additional control for genomic DNA contamination of the sample was assessed by including an RT-negative control for each RNA sample. The integrity of RNA was previously established with Affymetrix test chips showing acceptable 3′/5′ ratios for human glyceraldehyde-3-phosphate dehy-drogenase (GAPDH) gene < 2. The amount of cDNA in each sample was assessed by amplification of GAPDH using manufacturer’s primers and probes specific to the gene (ABI, TaqMan kit).
The reporter probes for each gene assayed were labeled with FAM (6-carboxyfluorescein) as the reporter and TAMRA (6-carboxy-4,7,2,7′-tetramethylrhodamine) as a quencher: XIST [5′(FAM)-TGAAGTAATAGATGTGAGATC-CAGACCGAAAGTCA-(TAMRA)3′], DBY [5′(FAM)-TCCT CAAACATGGTTATTTCTGTCAGTGACTTAACA-(TAMRA) 3′], SMCY [5′(FAM)-CCGATGCTCAGAAGTGTCTTGCCAG C-(TAMRA)3′], and RPS4Y [5′(FAM)-AGAGGCAAAGTA-CAAGTTGTGCAAAGTGAGG-(TAMRA)3′].
The forward and reverse PCR primers for XIST were 5′-AACCAGGAAAGAGCTAGTATGAGGAA-3′ and 5′-CATGG CCACTGTGGACTTTCT-3′, respectively. The forward and reverse PCR primers for DBY were 5′-GATTTTCAGTGATT GTCTGGTATATTTACA-3′ and 5′-TGCTGGCTGGTAAAA CCGA-3′, respectively. The forward and reverse PCR primers for SMCY were 5′-AGGTTGGTTCGTAAAGTCCA-CAC-3′ and 5′-GGAAATCACTCCTGTATGCTAGCA-3′, respectively. The forward and reverse PCR primers for RPS4Y were 5′-TGTTCACCGCATCACAGTG-3′ and 5′-ATTCCCTT-CACTCCCACAGTA-3′, respectively. The primers and probes for XIST, DBY and SMCY were designed in their last exons. A TaqMan probe for RPS4Y was designed to span the junction between exons 3 and 4.
Prior to set-up of the TaqMan RT-PCR assay, the primers were first used to verify that a single band on agarose gel was visualized of the proper amplicon size. Dilutions of human genomic DNA (male and female) were used to generate a standard curve for XIST, DBY and SMCY based upon the fluorescent signal present from each genomic DNA dilution. A cDNA clone for RPS4Y (GenBank BG714017; IMAGE 4796935) was used as a reference standard to assess the copy number. These primers and probe were designed to span the junction between exon 1 and exon 2.
Dilutions of human genomic DNA (male and female) were used to generate a standard curve based upon the fluorescent signal present from each genomic DNA dilution. Prior to set-up of the RT-PCR assay, the primers were first used to verify that a single band on agarose gel was visualized of the proper amplicon size. A cDNA clone for RPS4Y (GenBank BG714017; IMAGE 4796935) was used as a reference standard to assess the copy number since genomic DNA would give essentially zero amplification based upon primers and probe sets chosen.
To discriminate Y expression of genes in female and male brain by TaqMan PCR of the VCX family of genes we employed primers (Supplementary Method 2) that amplify the repeat regions of VCX-A, VCX-B, and VCX-B1. We designed TaqMan MGB probes to VCX-A, VCX-B, and VCX-B1 (Supplementary Method 2).
In addition to confirmation of microarray results with TaqMan probes, real-time PCR with SybrGreen dye was used to replicate the microarray results in human brain samples from an independent cohort of subjects (n = 6 males, n = 4 females; Table 1, lower half). Reverse transcription PCR was run on total RNA extracted from the 30 samples from DLPFC, anterior cingulate, and cerebellum in the second cohort. The cDNA was then used in a quantitative real-time PCR assay to confirm gene expression.
In Situ Hybridization Histochemistry
The right side of each brain was processed for in situ hybridization histochemistry using labeled oligo- or ribop-robes. Blocks were raised to 4ºC, fixed by immersion in 4% paraformaldehyde in 0.1 M phosphate buffer, infiltrated with sucrose and frozen in dry ice. Sections were cut at 25 μm and processed for radioactive in situ hybridization histochemistry. To generate subclones for riboprobe synthesis, the unique regions of h-SMCY (NCBI Genbank accession U52191, nucleotide coding 5072–5388), h-UTY (XM_034132, 2540–3036), and h-XIST-a (X56199, 567–952) from a human cDNA brain library (EdgeBiosystems, Gaithersburg, MD) were amplified using PCR. Amplified cDNA segments were extracted (QIAquick Gel Extraction Kit, Qiagen, Valencia, CA), subcloned (Zero Blunt TOPO PCR cloning kit, Invitrogen, Carlsbad, CA) and confirmed by nucleotide sequencing. Since some genes of interest did not differ sufficiently for optimal riboprobe design, oligonucleotide probes were synthesized for h-clone 23599 (NCBI Genbank accession #U79247, nucleotide coding region 1091–1106), h-USP9Y (AF000986, 1384–1412), h-RSP4Y (M58459), and h-DBY (AF000984, 3125–3157). 35S-or 33P-labeled sense and antisense oligo- or riboprobes were prepared and hybridized to sections from the DLPFC, anterior cingulate cortex, and cerebellum using previously published methods (Meador-Woodruff et al, 1989; Tighilet et al, 1998). Hybridized sections were exposed to Kodak XAR film for 2–4 weeks and after development of the film, the sections were dipped in Kodak autoradiographic emulsion and exposed for further 2–4 weeks, as described in the published methods.
RESULTS
All brain samples showed an acceptable level of RNA integrity based upon the ethidium bromide-stained gel analysis and the 28S/18S ribosomal bands which were generally greater than 1.5 for the cerebellar cortex samples (Table 2). The microarray quality control indicators of 3′/5′ ratios of GAPDH and β-actin were all less than 2.3. These values are within normal limits based on the manufacturer’s recommendations for fresh tissue or cell culture experiments.
Table 2.
Quality Control Indicators for Cohort I Samples used in Affymetrix Microarray and TaqMan PCR Analyses
| Pair | Gender | 3′/5′ β-actin | 3′/5′-GAPDH | Present call (%) | 28S/18S ratio |
|---|---|---|---|---|---|
| 1 | F | 2.22 | 2.15 | 30.7 | 2.22 |
| 2 | F | 1.92 | 1.36 | 33.9 | 1.92 |
| 3 | F | 2.16 | 1.59 | 32.2 | 2.16 |
| 4 | F | 2.62 | 2.23 | 32.0 | 2.62 |
| 5 | F | 1.44 | 1.34 | 37.9 | 1.44 |
| 1 | M | 1.53 | 1.3 | 37.0 | 1.53 |
| 2 | M | 2.06 | 1.64 | 33.1 | 2.06 |
| 3 | M | 2.07 | 1.96 | 33.2 | 2.07 |
| 4 | M | 1.79 | 1.81 | 32.2 | 1.79 |
| 5 | M | 1.84 | 1.94 | 29.1 | 1.84 |
The ratio of 3′/5′ for β-actin and GAPDH gene transcripts in cerebellum, present call percentage, and ratios of 28S/18S ribosomal RNA bands are shown for each sample.
Analysis of the microarray dataset was carried out using dCHIP software (Li and Wong, 2001). Significant differential expression between male and female brain samples was first evaluated, using criteria of greater than a 1.5-fold change (90% lower bound confidence interval limit) and a t-test value yielding a probability of p < 0.05. These criteria were chosen to minimize the false discovery of genes when three identical cRNA samples were split into technical duplicates, hybridized on separate chips, and the duplicate chips compared. A third criterion was established that each independent dCHIP analysis performed for each experimental replicate should replicate the same genes. When using only two criteria, that is, removing a 1.5-fold change requirement we did not, in fact, observe any additional genes that were significantly changed with the two remaining criteria.
Gender-Specific Gene Expression: Localization to Sex Chromosomes
We first present global gene expression differences between males and females combining all brain regions, and then secondarily look at regional differences by gender (Table 3). We identified six differentially expressed genes on sex chromosomes in male and female brain meeting three criteria of: 1.5-fold change, significant gender differences (t-test for gender differences, p < 0.05) and experimental replication across three sites independently running the microarray chips. Identification of five differentially expressed genes (XIST, DBY, SMCY, RPS4Y, USP9Y) was replicated in all three laboratories running the same gender samples. A sixth gene, UTY, was replicated at two sites only (Supplementary Table 1) but met the other two criteria. These global differences were seen when using a t-test based upon comparison of all male (n = 15) and female samples (n = 15) across three brain regions and replicated independently at three sites. When applying less stringent criteria, that is, no restriction on fold change but retaining a t-test significance of p<0.05, no additional genes met the criterion of replication across all laboratory sites. We repeated the same analysis using signal intensity generated with Affymetrix Microarray Suite software (MAS 5.0) and the same gene differences were found.
Table 3.
Fold Change of Differentially Expressed Sex Chromosome Genes Between Gender
| Gene | Fold changea | Female
|
Male
|
Affymetrix probeset | GenBank accession | ||
|---|---|---|---|---|---|---|---|
| Mean | SEM | Mean | SEM0 | ||||
| UTY | 2.1 (1.6) | 205.4 | 23.9 | 430.1 | 45.5 | 34477_at | AF000994 |
| USP9Y | 15.3 (5.1) | 37.4 | 40.9 | 571.7 | 88.5 | 35885_at | AF000986 |
| SMCY | 2.1 (1.6) | 487.7 | 47.8 | 1026.5 | 133.2 | 37583_at | U52191 |
| DBY | 78.7 (11.9) | 4.3 | 14.2 | 338.2 | 38.1 | 38355_at | AF000984 |
| RPS4Y | 127.8 (31.3) | 13.6 | 24.2 | 1737.4 | 249.2 | 41214_at | M58459 |
| XIST | −14.2 (−4.1) | 391.7 | 62.5 | 27.6 | 37.6 | 38446_at | X56199 |
The mean expression level averaged across three brain regions are shown for male and female groups.
The average fold change for male/female is shown in bold, and the fold change in parentheses is the estimated lower bound of the 90% confidence interval fold change. Data is shown for one independent analysis, but representative of all three sites. A negative number indicates that the female group was higher than the male group for XIST.
The expression of the six identified genes from the microarray data was analyzed for gender differences at the level of the three brain regions separately (Table 4). The six genes identified in the global analysis (vide supra) continued to show significant differences in expression in the Anterior Cingulate (AntCng) region comparisons (Table 4). The DLPFC and cerebellum regional comparisons also showed significant differences according to gender, but not at as high levels of significance, likely due to a reduction in the number of chips being compared, although trend level comparisons were exhibited for individual genes (p<0.12 for XIST, UTY, SMCY, and USP9Y in the DLPFC, and p<0.07 for SMCY in cerebellum). If we relaxed the criterion of fold change for determination of significant genes within a brain region by gender, there were no other consistent gene differences in our cohort of five males and five females brains that consistently met the criterion of experimental replication (p<0.05 by t-test) at three sites. The six genes consistently found in these sets of samples were localized to sex chromosomes, and no other genes were experimentally replicated at three sites.
Table 4.
Signal Intensity and Fold Change Summary Statistics for Six Sex Chromosome Genes by Region and Gender
| Region | Gene | Male | SEM | Female | SEM | FCa | p-Value |
|---|---|---|---|---|---|---|---|
| AntCng | XIST | 33 | 22 | 245 | 43 | −3.29 | 0.0018 |
| AntCng | DBY | 315 | 54 | −24b | 15 | 0.00 | 0.0002 |
| AntCng | RPS4Y | 1818 | 416 | 5 | 53 | 18.29 | 0.0019 |
| AntCng | SMCY | 1134 | 189 | 472 | 81 | 1.60 | 0.0105 |
| AntCng | USP9Y | 496 | 100 | −29b | 84 | 0.00 | 0.0030 |
| AntCng | UTY | 486 | 57 | 207 | 32 | 1.72 | 0.0021 |
| Cerebellum | XIST | 22 | 25 | 656 | 113 | −9.85 | 0.0004 |
| Cerebellum | DBY | 319 | 58 | 21 | 36 | 3.73 | 0.0018 |
| Cerebellum | RPS4Y | 1294 | 205 | 25 | 47 | 12.03 | 0.0002 |
| Cerebellum | SMCY | 789 | 85 | 525 | 98 | 1.08 | 0.0723 |
| Cerebellum | USP9Y | 583 | 82 | 76 | 73 | 2.84 | 0.0012 |
| Cerebellum | UTY | 364 | 41 | 202 | 24 | 1.37 | 0.0080 |
| DLPFC | XIST | 31 | 115 | 253 | 43 | −1.10 | 0.1043 |
| DLPFC | DBY | 371 | 95 | 11 | 20 | 7.48 | 0.0047 |
| DLPFC | RPS4Y | 2038 | 591 | 4 | 47 | 21.87 | 0.0075 |
| DLPFC | SMCY | 1128 | 348 | 445 | 61 | 1.22 | 0.0850 |
| DLPFC | USP9Y | 623 | 252 | 53 | 43 | 2.99 | 0.0524 |
| DLPFC | UTY | 465 | 120 | 242 | 57 | 1.02 | 0.1266 |
The average signal intensity is shown for male and female with the standard error of the mean (SEM).
FC is the ratio of fold change of male/female for each gene. The estimate of the 90% lower bound fold change calculated with dCHIP software (Li and Wong, 2001) is shown. Negative fold change indicates, as in Table 3, that gene expression is higher in female as compared to male brain.
The negative value for female gene expression (DBY, USP9Y) indicates that the dCHIP model estimates essentially zero expression.
Data shown for one site but representative of all three sites.
The top and bottom 1% of differentially expressed genes selected by fold change and t-test criteria only, were evaluated for over-representation in GO and KEGG classifications. The Y chromosome genes were over-represented in the DLPFC and cerebellum (p = 2.9 × 10−5 and 2.2 × 10−3, respectively, following a Bonferroni correction for multiple tests). There were no pathways or gene classifications that were significant in the anterior cingulate cortex following Bonferroni correction.
The regional microarray expression of XIST in females is variable between the cerebellum and cortical regions. The cerebellum showed an approximate 2.5 times greater XIST expression level compared with either cortical region. The regional coefficient of variation (CV) for XIST was 61% between the three regions for female samples. The CV for the Y-linked gene expression in males appeared considerably less than for the XIST gene. The CV of the five significant Y-linked genes in males across three brain regions was 8.6, 9.5, 13.4, 20.6, and 22.6% for DBY, USP9Y, UTY, SMCY, and RPS4Y, respectively. Thus, there appeared to be a more heterogeneous expression of XIST in female brain compared to the five Y-linked genes in the same brain regions of males.
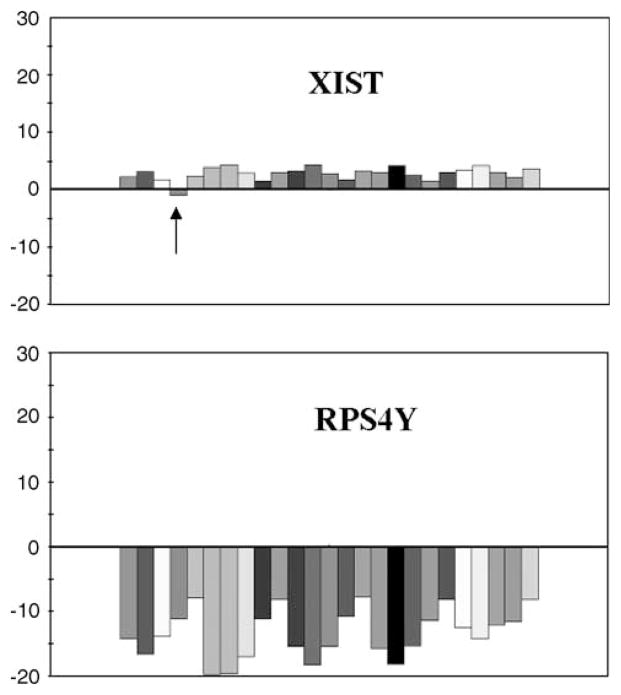
The consistency of the DLPFC findings was further examined by computing the ratio value for each possible male and female gene comparison using Affymetrix Microarray Suite (software version 4.0). A total of 150 combinations of comparisons were made (six genes × five males × five females). In 149 of 150 comparisons the direction of the fold change was consistent with upregula-tion in male samples compared to female for the Y-linked genes and the reverse for the X-linked gene XIST (Figure 1).
Figure 1.
The fold change of gene expression for five males and five females samples from the DLPFC. (XIST) A predominant female expression of XIST (an exception is noted with an arrow) is shown. A higher ratio for female is shown as positive above zero, and lower expression for female is shown as below zero. (RPS4Y) The pattern for RPS4Y gene shows a predominant male expression. Both graphs depict each female sample/male combinations for five female and five male samples. The Y-axis scale ranges from −30- to 30-fold change and is calculated for each individual female/male combination in Affymetrix Microarray Suite version 4.0.1.
The data was also analyzed by the present and absent calls parameters generated for the DLPFC in Affymetrix Micro-array Suite software. There was a range of 60–100% present calls for the five Y-linked genes in male (RPS4Y, UTY, SMCY, DBY, USP9Y) and 0% present calls in females for these Y-linked genes across all three brain regions. For XIST, the present calls were 95% for female samples and 0% present calls for male samples across all brain regions. The gender classification of each sample was accurately predicted by subtracting the microarray expression of DBY from XIST, which reliably differentiated male and female samples in all brain regions.
There were 26 probe sets that represented genes found on the Y chromosome on the Affymetrix HGU95A version 2 microarray. In brain, this Affymetrix Microarray was able to detect five significant Y genes. The Y-linked genes are expected to be found only in male brain. The remaining 21 genes showed a level of approximately equal expression in male and female samples, although expression was at background levels for some Y chromosome genes. We questioned why there were not differences between male and female brain samples for these 21 Y-linked genes. The lack of differences most likely result from the following: (1) a lack of probe set specificity, since Y chromosome sequences also are found in autosomal genes, or are sufficiently similar to X homologues to cross-hybridize to the Y probe set (Skaletsky et al, 2003), (2) a background level of expression by microarray, that is, not present in brain or expression levels are below the detectable limits of microarray sensitivity, and (3) the probe sets gave low present calls despite an above background reading on the microarray due to singular probe-level cross-hybridization. Thus, Y-linked genes that appear to be found in female brain result from the nature of the hybridization probes and suggest that careful attention to design and interpretation of results using Affymetrix probe sets is warranted. We examine the issue of probe set cross-hybridization further in the technical section (vide infra).
Homologous X-Chromosome Genes
We elected to look at the X homologues (DBX, UTX, SMCX, RPS4X) for the differentially expressed Y chromosome genes for possible differences between genders. XIST does not have a homologue on the Y chromosome. We expect that these X homologues (DBX, UTX, SMCX, RPS4X) might be differentially expressed since they escape X inactivation. However, these X homologues were expressed at comparable levels in male and female brain samples (Supplementary Table 2). We re-examined the entire set of X-chromosome gene probe sets by normalizing each Affyme-trix chip to the 50th percentile signal across all chips. Only the XIST gene was differentially expressed as reported above (Supplementary Result, #2; Supplementary Data Table 3).
Real-Time PCR Validation of Differentially Expressed Genes
To validate the set of gender genes, we examined RPS4Y, SMCY, DBY, and XIST genes by TaqMan real-time PCR (Table 1, Cohort 1) and SybrGreen real-time PCR (Table 2, Cohort 2). Both real-time methods and cohorts yielded the same results and conclusions. The gene copy number mean ( ± SD) for TaqMan real-time PCR results show a significant difference between males and females in the expression of these genes (RPS4Y, SMCY, DBY, and XIST) in the DLFPC (t-test p-values range from 0.002 to 0.0007, Table 5). The TaqMan real-time PCR fold change results exceeded the fold changes observed by microarray (Table 5) with good agreement between log intensity values for TaqMan and microarray (r = 0.76).
Table 5.
Sex Chromosome Genes Assayed with TaqMan Real-Time PCR in Dorsolateral Prefrontal Cortex
| Gender | Gene copy mean (SD)
|
|||
|---|---|---|---|---|
| RPS4Ya,b | SMCYb | DBYb | XISTb | |
| Female | 33 (28) | −506 (320) | 5.9 (8.0) | 34 640 (8297) |
| Male | 53 200 (21 370) | 9813 (3208) | 4002 (820) | 281 (97) |
| t-testc | 0.005 | 0.002 | 0.002 | 0.0007 |
The mean gene copy number (SD) are shown for RPS4Y, SMCY, DBY, and XIST in male and female samples.
The RPS4Y clone was also used as a reference standard in the assay for copy number, and values for male 36 000 (15 427) and female 30.8 (19.3) with the same results as genomic DNA standard as shown in the table. The RT− sample yielded a negligible copy number of 0.2 and 0.8 for male and female mean.
Genomic DNA standard with RT− sample copy number subtracted from RT+ copy number. A negative value indicates that genomic presence in total RNA sample gives a reading higher than the RT+ sample.
t-Test p-values unequal variance between groups.
To determine whether these gene expression differences would replicate in an independent cohort we examined additional RNA samples from controls (n = 6 males and n = 4 females; shown in Table 1, lower half) from the DLPFC, anterior cingulate, and cerebellum, with real-time PCR. We consistently found the same results for the five differentially expressed Y-linked genes with real-time SybrGreen PCR. The results of real-time PCR with SybrGreen showed PCR cycle differences ranging from 9 to 12 between male and female samples for DBY, RPS4Y, SMCY, USP9Y, and UTY depending on the brain region and gene. The t-test comparisons between males and females using real-time PCR SybrGreen data were significant for each brain region (two-tailed Student’s t-test p-values range from 0.02 to 0.00001 for 15 t-tests). The real-time PCR validation of the five Y genes also showed differential expression by region. Thus, the real-time TaqMan and SybrGreen PCR verification of the gender genes confirm the microarray data and support the differential expression of these sex chromosome genes in three brain regions examined (Table 5).
In Situ Hybridization Histochemistry
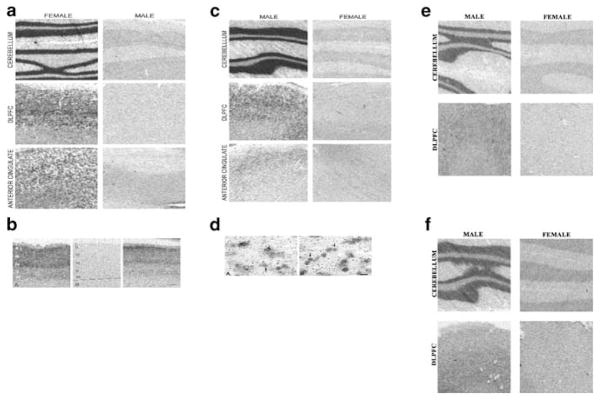
The sex chromosome genes have not been systematically studied in human brain. We elected to understand the nature of the differential expression by using in situ hybridization histochemistry to address possible cellular and layer-specific localization of the genes in three brain regions. In situ hybridization with probes specific for XIST, UTY, SMCY, and RPS4Y produced strong signal in a gender-specific manner as expected from the microarray and real-time PCR data. The XIST riboprobe showed clear labeling only in female brains with a laminar-specific distribution in the DLPFC (Figure 3). XIST hybridization signal was present in all sections from the DLPFC of females, but absent in all sections from males. Male brains in all three regions showed only background labeling with the XIST riboprobe (Figure 3a, b) similar to the background levels for the negative control using XIST riboprobe on an RNase-treated section. In female cortex, the XIST riboprobe label was stronger in superficial cellular layers (II–IV) than in deeper layers (V–VI). Labeling of layer I and the white matter was at background levels. XIST riboprobe labeling of most granule cells and Purkinje cells was strong in the cerebellum. The XIST laminar distribution in the cortex and cerebellum and the lack of hybridization in white matter imply that XIST is prevalently expressed in neurons. Further silver grain analysis of emulsion dipped slides confirms that predominantly pyramidal cells in the DLPFC are labeled with XIST riboprobe and there was a lack of silver grains over Nissl-stained somata of neuroglial cells (Figure 3c). Silver grain analysis of emulsion dipped cerebellar slides confirmed that granule and Purkinje neurons in cerebellum are also labeled. The XIST in situ hybridization histochemistry was repeated with two different methods (Gee et al, 1983; Tighilet et al, 1998) and gave similar conclusions.
Figure 3.
In situ hybridization of male and female brain sections from the cerebellum, DLPFC, and anterior cingulate cortex. (a) The XIST antisense probe is shown for six sections (three males and three females) with significant pattern of labeling in neurons from female sections. (b) The XIST probe shows female DLPFC hybridization signal (left and right panels) was present in all layers of cortex with laminar-specific distribution and absent in male DLPFC layers (middle panel). In general signal was stronger in superficial layers than in deeper layers for female DLPFC. The laminar distribution and lack of hybridization in white matter suggest that XIST is prevalently expressed in neurons. (c) The 35S-XIST riboprobe silver grains reveals dense labeling of neurons and very sparse and usually absent labeling of glia. (d) The RPS4Y probe shows predominant pattern of labeling in neurons for male tissues. (e) The SMCY probe and UTY probe (f) also confirms the microarray and TaqMan/real-time PCR experiments showing a strong labeling of male cerebellum, which shows higher tissue labeling compared to both cortical regions.
There was qualitative correspondence between in situ hybridization histochemistry and microarray abundance in different brain regions. For example, there were lower levels by in situ hybridization histochemistry signal in cortical regions for SMCY and UTY (Figure 3d, e) compared to XIST and RPS4Y cerebellar levels (Figure 3a, b) that correspond to relative differences observed by microarray. The in situ hybridization histochemistry signal for the SMCY and UTY sections (Figure 3d, e) also took significantly longer film exposure time to visualize as compared to XIST and RPS4Y. Although further analyses at the silver grain level are required, similar to the XIST gene, the Y-linked genes (SMCY, UTY, RPS4Y) appear to be localized to neurons in the cerebellar and cortical regions as determined with in situ hybridization histochemistry. The roles of these neuronal genes in neurobiology will be of interest, especially since it appears that not all neurons express these gender-specific genes in cortical and cerebellar regions.
Technical Issues Involved in Microarray Detection of Genes
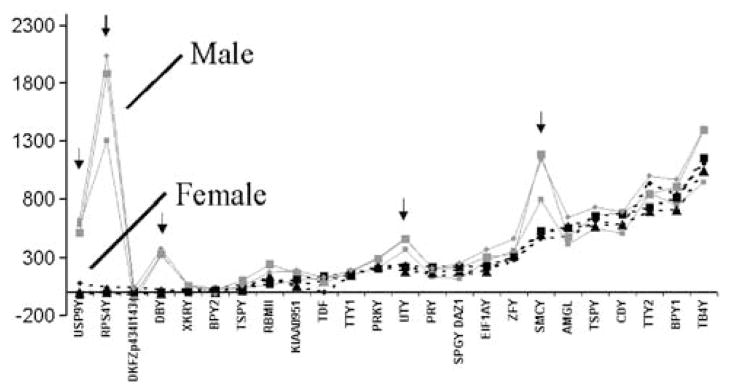
We found that certain Y chromosome genes were apparently expressed at equivalent levels in female and male brain tissue (Figure 2). In this section, we explore the possible reasons for detection of Y chromosome genes in female brain with subsequent experiments and analysis. The microarray technical issues in turn can help to further understand the biological findings.
Figure 2.
The mean gene expression level (y-axis) of the Y-linked probesets shown for three brain regions (large square represents anterior cingulate cortex, circle represents DLPFC, small square represents cerebellum) for male (red-solid line) and female (black-dashed line) subjects. The gene order on the x-axis is first sorted from lowest gene expression in female samples to the highest. We found consistent differences in Y expression of five genes denoted by arrows (RPS4Y, USP9Y, DBY, UTY, SMCY). We confirmed that the BPY1 gene showing above background expression in female and male brain tissue is a result of an X-homologue (VCX) expression in brain tissue. Another corresponding gene BPY2 was absent for both male and female brain tissue and confirmed to be absent by RT-PCR. Cross-hybridization of autosomal and X-linked genes with the Y probeset is a probable cause for the high expression of female brain tissue for Y-linked genes (see Table 6 for additional information).
TaqMan RT-PCR Validation of Testis-Specific Genes
We chose the VCY gene as an example of anomalous Y gene expression, which appeared to be highly expressed in female and male brain tissue (Figure 2). The VCX genes appear to be expressed in the brain according to microarray results and were further studied by TaqMan RT-PCR to determine the specificity of the microarray findings. The VCX, VCY (BPY), and VCY2 (BPY2) genes were analyzed by RT-PCR for expression in brain and testis. We designed specific primers for VCX, VCY, and VCY2 genes (Supplementary Method 2). Further we utilized a MGB nonfluorescent quencher probe for discrimination of single base-pair differences between the VCX and VCY/VCY2 genes. Our TaqMan RT-PCR MGB results showed that VCY2 was expressed in testis (Supplementary Table 4) and not expressed in brain tissue from the male or female samples of DLPFC, which is consistent with the microarray finding. Further the VCY gene (BPY1) was not expressed in brain in either male or female DLPFC by TaqMan RT-PCR contrary to the microarray result that shows abundant expression in male and female brain. We could detect VCX in low abundance in DLPFC by RT-PCR (Supplementary Table 4) and there were no expression differences between genders for VCX. These results indicate that the high microarray expression of VCY in both male and female samples is most likely due to a cross-hybridization from the VCX mRNA with the VCY probe set in brain. The absence of VCY2 expression in brain could not contribute to the spurious finding of VCY expression in male and female brain.
We next asked whether the Y chromosome genes on the Affymetrix chip would be specific in recognition of testicular or ovarian tissue total RNA. If the elevated levels of Y-linked genes are due to cross-hybridization artifacts in female samples, then it would be reasonable to ask if the same Y-linked genes would be also elevated in ovarian tissue samples. A database of human ovary and testes samples run on HGU95A chips was queried (http://expression.gnf.org/cgi-bin/index.cgi) at Novartis Gene Expression (Su et al, 2002). The query consisted of all Y probe sets on the Affymetrix chip and examining the average expression levels for these Y chromosome probe sets in ovarian and testicular tissues. Probe set specificity was defined as the ratio of gene expression in testis/ovary and an arbitrary ratio greater than 2 indicated probe set specificity. The data (Table 6) shows clearly that 10 probe sets are specific for detection of male transcripts in testes but not ovary. Five Y chromosome probe sets do not differentiate between testis and ovary (arbitrary ratio <2) thus indicating that the equivalent levels of these five probe sets in brain samples from male and female are due to a lack of probe set specificity (For comparison, see Figure 2). We cannot specifically address issues regarding probe sets that did not show any levels in either ovary or testes. The overall results of this examination of the Y chromosome gene expression in ovary and female brain shows that when microarray differences in Y chromosome expression were evident between males and females, it was always accompanied by specificity of the probe set defined as a high testis/ovary ratio. Conversely, there were no examples of significant Y gene expression differences between males and females when there was a lack of probe set specificity for Y gene expression.
Table 6.
Comparison of Y Chromosome Probesets in Male and Female Brain Tissue to Testicular and Ovarian Expression Levels
| Gene symbol | Testis/ovary | M/F brain | Probe discriminates |
|---|---|---|---|
| TSPYQ1 | 36.58 | 2.84 | Yes |
| RPS4Y* | 32.41 | 79.90 | Yes |
| DBY* | 23.12 | 6.64 | Yes |
| SMCY* | 11.78 | 2.07 | Yes |
| VCY | 10.77 | 1.12 | Yes |
| DAZ | 7.93 | 1.02 | Yes |
| ZEY | 6.70 | 1.27 | Yes |
| UTY* | 3.70 | 2.09 | Yes |
| EIF1AY | 3.68 | 1.59 | Yes |
| USP9Y* | 2.76 | 183.37 | Yes |
| PRKY | 1.84 | 1.24 | No |
| TTTY2 | 1.47 | 1.16 | No |
| TTTY15 | 1.25 | 3.25 | No |
| None | 1.20 | 1.52 | No |
| TMSB4Y | 1.11 | 1.13 | No |
| SRY | 1.00 | 12.52 | Not expressed |
| XKRY | 1.00 | 5.42 | Not expressed |
| KIAA0951 | 1.00 | 2.22 | Not expressed |
| VCY2 | 1.00 | 1.83 | Not expressed |
| PRY | 1.00 | 1.00 | Not expressed |
| None | 0.70 | 1.06 | No |
| TTTY1 | 0.65 | 1.15 | No |
| AMELY | 0.52 | 0.96 | No |
| CDY1 | 0.44 | 1.02 | No |
Gene symbols shown with * were found to be differentially expressed by microarray analysis of male and female brain samples. The M/F brain ratio is an average of three brain region ratios for male and female gene expression. Testis and ovary expression levels were obtained from Novartis website http://expression.gnf.org. A ratio of testis/ovary expression greater than 2 was arbitrarily set to discriminant between tissues. A ratio of testis/ovary expression of 1 indicates the probeset was at background level for both the testis and ovary samples. In each case where brain microarrray differences were found (*), there was a ratio greater than 2 for discrimination between ovarian and testicular mRNA samples for the same genes.
DISCUSSION
We first address the biological implications of the microarray findings that were validated by RT-PCR and in situ hybridization histochemistry. The technical issues that are applicable to microarray studies designed similar to the present study are then discussed. We have identified problems due to cross reactivity of genes that bear great sequence identity.
However, it does appear that we can also identify a subset of X and Y sex chromosome genes that behave as expected using multiple assays. Moreover, the pattern of expression of these genes in the brain is intrinsically very interesting and leads to further biological questions.
Sex Chromosome Linked Gene Expression
Gender-specific gene expression in brain
The possible neurobiological impact of the six differentially expressed X-and Y-linked genes are important since these genes have varying sequences in males and females. A dimorphic expression pattern could translate into functional differences. Further, the genes appear to be preferentially expressed in subsets of neurons. For the XIST gene there is no homologue on the Y chromosome, so XIST mRNA is uniquely expressed in female tissue and inactivates a long stretch of genes near the centromere of the X chromosome (Brockdorff et al, 1991). In fact, XIST does not appear to encode a protein and probably exerts modulatory influence over the X chromosome as a structural RNA within the nucleus (Brown et al, 1992). The present study confirms that XIST mRNA is expressed only in female brain and further demonstrates that it is only expressed in neurons. XIST may coordinate gene expression in specific neuron populations. Since XIST is expressed in only a subset of neurons in three brain regions, it is tempting to speculate about the potential neuronal phenotypes that are generated between XIST+ and XIST− neuronal cells. XIST expression is heterogenous within a brain region, that is, a large CV is shown between individual samples within a brain region. An intriguing question raised by our observations is whether XIST+ neurons silence gene expression causing functional differences compared to XIST− neurons. Gene products of the sex chromosomes are thought to regulate differences in the number of midbrain dopaminergic neurons in males and females produced in vitro (Carruth et al, 2002; Sibug et al, 1996) independent of steroidal control by gonadal tissue (Carruth et al, 2002). The number of dopaminergic neurons can be altered by sex chromosome gene expression, and thus vulnerability to disorders involving midbrain dopamine neurons might be manipulated through similar functional pathways.
The DEAD box protein family includes DBX and DBY and share asp-glu-ala-asp amino-acid residues represented by the amino-acid letters D-E-A-D. DEAD box proteins are implicated in splicing, ribosome assembly, translation initiation, spermatogenesis, and embryogenesis. The possible differences between DBY and DBX have been described (Lahn and Page, 1997). DBX is one of five X-linked genes that have homologues located in the nonrecombining region of the Y chromosome (Lahn and Page, 1997). It is postulated that DBX and four other genes that escape X inactivation are cases in which gene expression is maintained at comparable levels in males and females by preservation of homologous genes on both the X and the nonrecombining region of the Y chromosome, with male and female cells expressing both copies of each gene (Lahn and Page, 1997). The DBY gene encodes a MHC Class II antigen that is recognized by lymphocytes and involved in graft rejection. The DBX protein was not recognized by lymphocytes (Vogt et al, 2002). DBY is expressed ubiquitously in male tissues.
Both RPS4Y and RPS4X encode ribosomal protein S4. Fisher et al (1990) demonstrated that the RPS4Y and RPS4X proteins differ at 19 of 263 amino acids. Since these genes are ubiquitously expressed in human tissues, it was suggested that the ribosome of human males and females might have structurally distinct proteins. RPS4X is not dosage compensated, and is known to escape X inactivation (Fisher et al, 1990). The present microarray results confirm that RPS4X is found in equal levels between normal male and female brain samples, which may be due to the close similarity between RPS4X and RPS4Y genes (Skaletsky et al, 2003).
The functions of SMCY, USP9Y, RPS4Y, DBY, and UTY can be inferred from animal studies as no human brain studies have been reported to our knowledge. The five Y genes we studied may play roles in the brain and contribute to behavioral differences observed in animal studies, such as aggressive behavior in mice, hippocampal size asymmetry and mossy fiber distribution, and discrimination learning (Xu et al, 2002). Interestingly, four genes were localized in neurons (SMCY, UTY, RPS4Y, XIST) as determined with in situ hybridization histochemistry. The roles of these neuronal genes in neurobiology will be of interest, especially since it appears that a subset of neurons express these gender-specific genes. A prior report of gene expression of ZFY and SRY in adult human cortex (Mayer et al, 1998) commented that male-specific transcriptional regulation in neurons might occur as a result of the expression of these two DNA binding proteins, ZFY and SRY:
“We propose therefore that they function as sex-specific cell-intrinsic signals that are needed for full differentiation of a male human brain, and that continuous expression throughout life may be required to maintain sex-specific structural or functional properties of differentiated male neurons” (Mayer et al, 1998, p 285).
The expression in our microarray data was low for SRY (TDY shown in Figure 2), but did show a male to female ratio of 12.5, in line with the prior report of low abundance in male cortex and absence in female cortex (Mayer et al, 1998). Moreover, the abundance of SRY protein expression was too low to measure by immunohistochemistry in postmortem human brain (Mayer et al, 1998). For ZFY, a modest difference between male and female brain samples was found by microarray analysis (ratio of 1.27) that was not statistically significant. The lack of differences between genders in the ZFY gene is most likely due to 95% homology to the ZFX homologue (Mayer et al, 1998).
Overall, the functional role of the X and Y sex chromosome gene pairs in male brain is not known. Each of the differentially expressed Y genes have a closely related X homologue. Some of these X–Y homologues are thought to arise from an original autosomal chromosome. Parts of the X and Y segments of the sex chromosomes no longer recombine in males, which have allowed the X–Y gene pairs to evolve separately (Graves, 1995). However, whether each of the X–Y homologues is functionally interchangeable in brain function has not been determined. Separate functions for the Y genes in male sexual differentiation, such as spermatogenesis, are known. In brain, the functional differences due to expression of a Y chromosome gene could be diminished by the expression of its X partner. While speculative, the differences in the five Y chromosome genes expressed in brain might be responsible for subtle changes in brain function and morphology between males and females. Adding the prior report of ZFY and SRY expression in human brain, we have seven Y chromosome genes expressed in human brain that may regulate gender differences.
X chromosome gene expression
In the present study of specific human brain regions the X homologues (DBX, UTX, USP9X, RPS4X, and SMCX) of the five Y genes show the same expression in males and females. However, we cannot conclusively state that the levels in human male and female brain are equivalent for the X homologues, due to the possibility of cross-hybridization. Gender differences were reported in mouse brain gene expression for Usp9y, Ube1y, Smcy, Eif2s3y, Uty, and Dby (Xu et al, 2002) and in fetal mouse brain corresponding to the Xist gene and the male-specific Smcy gene (Eriksson et al, 1999). The X homologues (Usp9x, Ube1x, Smcx, Utx, and Dbx) were expressed approximately two-fold higher in whole-brain homogenates from female mice compared to male mice (Xu et al, 2002). In our human data we are drawn to the opposite conclusion that the sum of the X+Y homologue is increased in males due to the equal X-homologue levels between males and females (Supplementary Table 2), and the absence of Y expression for these five genes in females. A recent study using a focused X-linked gene microarray found that normal male and female lymphoblastoid cell lines differed only in the XIST gene expression in 100% of the microarray assays. In the same microarray analysis, no other specific gene differences among 1317 X-chromosome genes assayed were reported (Sudbrak et al, 2001). Thus, we cannot presently determine the exact reason for the lack of increased X-gene expression in human female brain beyond probe set specificity (Supplementary Discussion 1).
Technical Issues Involved in Microarray Detection of Genes
Cross-hybridization of probe sets
The factor that could account for the similar expression levels of the X homologues (DBX–DBY), (USP9X–USP9Y), (UTX–UTY), (RPS4X–RPS4Y) is the cross-hybridization among the probe sets chosen to represent the genes on the Affymetrix U95Av2 chip. One gene pair, USP9Y–USP9X, does not appear to show cross-hybridization in the probe set alignments. DBX–DBY probe sets overlap in 108 nucleotides over a 128 nucleotide stretch on the Affymetrix chip. Also, cross-hybridization between autosomal genes and Y and X probe sets were found. For example, one predicted ENSEMBL gene (ENST00000270291) is found on chromosome 19 and is matched to three probes for the RPS4X gene. There is also close similarity between RPS4X and RPS4Y genes for additional cross-hybridization signal (Skaletsky et al, 2003).
By designing specific PCR primers to Y chromosome genes we could find differential expression for five Y genes in males. Seven additional Y genes are testis specific, and expression of additional Y genes in the brain was below background detection. Five Y genes (AMLY, TB4Y, TSPY, CDY, VCY) give rise to signal in male brain equivalent to expression for female brain tissues. These probe sets should theoretically show no expression of a Y gene in female tissue. We found two of these genes were infrequently called present (TSPY (3%), CDY (3%)) in any sample regardless of gender, so that the actual detection might not be deemed reliable. Two of the Y-linked genes (AMLY and TB4Y) have overall homology to X-linked genes (AMELX (88%), and TB4X (93%)). Another Y gene, TSPY, is homologous with an autosomal gene TSPYL (Unigene Hs.278479) in two stretches (75–78% homology) and TSPYL is highly represented in SAGE cortical libraries. We demonstrated expression of the fifth gene (VCY) in female brain was presumably due to VCX expression by a sensitive TaqMan MGB probe in a real-time PCR assay. Finally, using data from the Novartis website (http://expression.gnf.org/cgi-bin/index.cgi) (Su et al, 2002), we were also able to show these five Y probe sets (AMLY, TB4Y, TSPY, CDY, VCY) also do not differentiate between male testes and female ovary tissue samples (Table 6). Clearly the five Y genes that we found different in human brain between males and females successfully differentiate testes from ovary gene expression (Table 6). Thus, cross-hybridization of probes on the Affymetrix Microarray must be considered in evaluation of the results.
SUMMARY
We have focused our study of gender differences in gene expression on the set of genes found on the sex chromosomes. We found differences in gene expression between genders in human brain for six sex chromosome genes (DBY, SMCY, UTY, RPS4Y, USP9Y, XIST). These six genes were also validated with real-time PCR. Interestingly, four of the six genes were localized in neurons (SMCY, UTY, RPS4Y, XIST) as determined with in situ hybridization histochemistry. The roles of these neuronal genes in neurobiology will be of interest, especially since it appears that subsets of neurons express these gender-specific genes. For example, XIST can silence gene expression on part of the X chromosome in females and is localized to a subset of neurons. Differential expression of genes from the sex chromosome might influence the development of mono-aminergic neurons (Reisert and Pilgrim, 1991). As an example, alterations in the number and sensitivity of midbrain dopaminergic neurons could result from the expression of the sex chromosome genes in the brain (Carruth et al, 2002).
Microarray technology is likely to extend our understanding of male and female differences in gene expression, but will require probe set refinement as shown by expression of Y-linked probe sets in female tissue. With further refinements in the Y chromosome sequence available (Skaletsky et al, 2003), probe sets can be redesigned to overcome some of the technical issues identified in this study. As we continue to examine neuropsychiatric disorders, we will build a database of control gene expression from a much larger number of brains. By examining restricted age ranges in future studies, this will answer developmental gender differences in brain maturation and aging. Fundamental biological questions concerning the role of the X and Y chromosomes in genetic determination of sex differences in brain function will require additional studies. The expression of the sex chromosome genes may contribute to gender differences in brain function independent of gonadal steroid influences and ultimately give rise to neuronal subsets with dimorphic patterns of gene expression and function.
Supplementary Material
Acknowledgments
We appreciate the assistance of Richard Stein, PhD, David Walsh, PsyD, and Preston Cartagena, PsyD for their contributions to post-mortem clinical characterization of subjects and Kathleen Burke for procurement of brain tissue, as well as Jacque Berndt and the investigators and medical examiners at the Orange County Coroners Office. We also appreciate the technical contributions of Erick Ferran, Karen Lopez, Sharon Burke, and Phong Nguyen. F Warren Lovell, MD, performed a neuropathological evaluation of the post-mortem brains. Tissue specimens were processed and stored at the Human Brain and Spinal Fluid Resource Center, Veterans Medical Center, Los Angeles under the direction of Wallace W Tourtellotte, MD, PhD. This project is supported by the NIMH Conte Center Grant P50 MH60398, Pritzker Neuropsychiatric Disorders Research Consortium, William Lion Penzner Foundation (UCI), Della Martin Foundation (UCI), NIMH Grant #MH54844 (EGJ), WM Keck Foundation (EGJ), and the NIMH Program Project MH42251 (SJW and HA).
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4. American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- Andreasen NC, Flaum M, Swayze V, II, O’Leary DS, Alliger R, Cohen G, et al. Intelligence and brain structure in normal individuals. Am J Psychiatry. 1993;150:130–134. doi: 10.1176/ajp.150.1.130. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Carruth LL, Reisert I, Arnold AP. Sex chromosome genes directly affect brain sexual differentiation. Nat Neurosci. 2002;5:933–934. doi: 10.1038/nn922. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:1–11. Article R60. [PubMed] [Google Scholar]
- Eriksson A, Wahlestedt C, Nordqvist K. Isolation of sex-specific cDNAs from fetal mouse brain using mRNA differential display and representational difference analysis. Brain Res Mol Brain Res. 1999;74:91–97. doi: 10.1016/s0169-328x(99)00265-x. [DOI] [PubMed] [Google Scholar]
- Escalona PR, McDonald WM, Doraiswamy PM, Boyko OB, Husain MM, Figiel GS, et al. In vivo stereological assessment of human cerebellar volume: effects of gender and age. AJNR Am J Neuroradiol. 1991;12:927–929. [PMC free article] [PubMed] [Google Scholar]
- Fisher EM, Beer-Romero P, Brown LG, Ridley A, McNeil JA, Lawrence JB, et al. Homologous ribosomal protein genes on the human X and Y chromosomes: escape from X inactivation and possible implications for Turner syndrome. Cell. 1990;63:1205–1218. doi: 10.1016/0092-8674(90)90416-c. [DOI] [PubMed] [Google Scholar]
- Gee CE, Chen CL, Roberts JL, Thompson R, Watson SJ. Identification of proopiomelanocortin neurones in rat hypothalamus by in situ cDNA-mRNA hybridization. Nature. 1983;306:374–376. doi: 10.1038/306374a0. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Snell JW, Lange N, Rajapakse JC, Casey BJ, Kozuch PL, et al. Quantitative magnetic resonance imaging of human brain development: ages 4–18. Cereb Cortex. 1996;6:551–560. doi: 10.1093/cercor/6.4.551. [DOI] [PubMed] [Google Scholar]
- Graves JA. The evolution of mammalian sex chromosomes and the origin of sex determining genes. Philos Trans R Soc Lond B Biol Sci. 1995;350:305–311. doi: 10.1098/rstb.1995.0166. discussion 311–312. [DOI] [PubMed] [Google Scholar]
- Gur RC, Mozley PD, Resnick SM, Gottlieb GL, Kohn M, Zimmer-man R, et al. Gender differences in age effect on brain atrophy measured by magnetic resonance imaging. Proc Natl Acad Sci USA. 1991;88:2845–2849. doi: 10.1073/pnas.88.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EG, Hendry SH, Liu XB, Hodgins S, Potkin SG, Tourtellotte WW. A method for fixation of previously fresh-frozen human adult and fetal brains that preserves histological quality and immunoreactivity. J Neurosci Methods. 1992;44:133–144. doi: 10.1016/0165-0270(92)90006-y. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lahr G, Swaab DF, Pilgrim C, Reisert I. The Y-chromosomal genes SRY and ZFY are transcribed in adult human brain. Neurogenetics. 1998;1:281–288. doi: 10.1007/s100480050042. [DOI] [PubMed] [Google Scholar]
- Meador-Woodruff JH, Mansour A, Bunzow JR, Van Tol HH, Watson SJ, Jr, Civelli O. Distribution of D2 dopamine receptor mRNA in rat brain. Proc Natl Acad Sci USA. 1989;86:7625–7628. doi: 10.1073/pnas.86.19.7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakkenberg B, Pelvig D, Marner L, Bundgaard MJ, Gundersen HJ, Nyengaard JR, et al. Aging and the human neocortex. Exp Gerontol. 2003;38:95–99. doi: 10.1016/s0531-5565(02)00151-1. [DOI] [PubMed] [Google Scholar]
- Rabinowicz T, Petetot JM, Gartside PS, Sheyn D, Sheyn T, de CM. Structure of the cerebral cortex in men and women. J Neuropathol Exp Neurol. 2002;61:46–57. doi: 10.1093/jnen/61.1.46. [DOI] [PubMed] [Google Scholar]
- Reisert I, Pilgrim C. Sexual differentiation of monoaminergic neurons—genetic or epigenetic? Trends Neurosci. 1991;14:468–473. doi: 10.1016/0166-2236(91)90047-x. [DOI] [PubMed] [Google Scholar]
- Sibug R, Kuppers E, Beyer C, Maxson SC, Pilgrim C, Reisert I. Genotype-dependent sex differentiation of dopaminergic neurons in primary cultures of embryonic mouse brain. Brain Res Dev Brain Res. 1996;93:136–142. doi: 10.1016/0165-3806(96)00024-7. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Su A, Cooke M, Ching K, Hakak Y, Walker J, Wiltshire T, et al. Large-scale analysis of the human and mouse transcrip-tomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbrak R, Wieczorek G, Nuber UA, Mann W, Kirchner R, Erdogan F, et al. X chromosome-specific cDNA arrays: identification of genes that escape from X-inactivation and other applications. Hum Mol Genet. 2001;10:77–83. doi: 10.1093/hmg/10.1.77. [DOI] [PubMed] [Google Scholar]
- Tighilet B, Huntsman MM, Hashikawa T, Murray KD, Isackson PJ, Jones EG. Cell-specific expression of type II calcium/calmodulin-dependent protein kinase isoforms and glutamate receptors in normal and visually deprived lateral geniculate nucleus of monkeys. J Comp Neurol. 1998;390:278–296. doi: 10.1002/(sici)1096-9861(19980112)390:2<278::aid-cne10>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Vogt MH, van den Muijsenberg JW, Goulmy E, Spierings E, Kluck P, Kester MG, et al. The DBY gene codes for an HLA-DQ5-restricted human male-specific minor histocompatibility antigen involved in graft-versus-host disease. Blood. 2002;99:3027–3032. doi: 10.1182/blood.v99.8.3027. [DOI] [PubMed] [Google Scholar]
- Xu J, Burgoyne PS, Arnold AP. Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet. 2002;11:1409–1419. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- Xu J, Kobayashi S, Yamaguchi S, Iijima K, Okada K, Yamashita K. Gender effects on age-related changes in brain structure. AJNR Am J Neuroradiol. 2000;21:112–118. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.