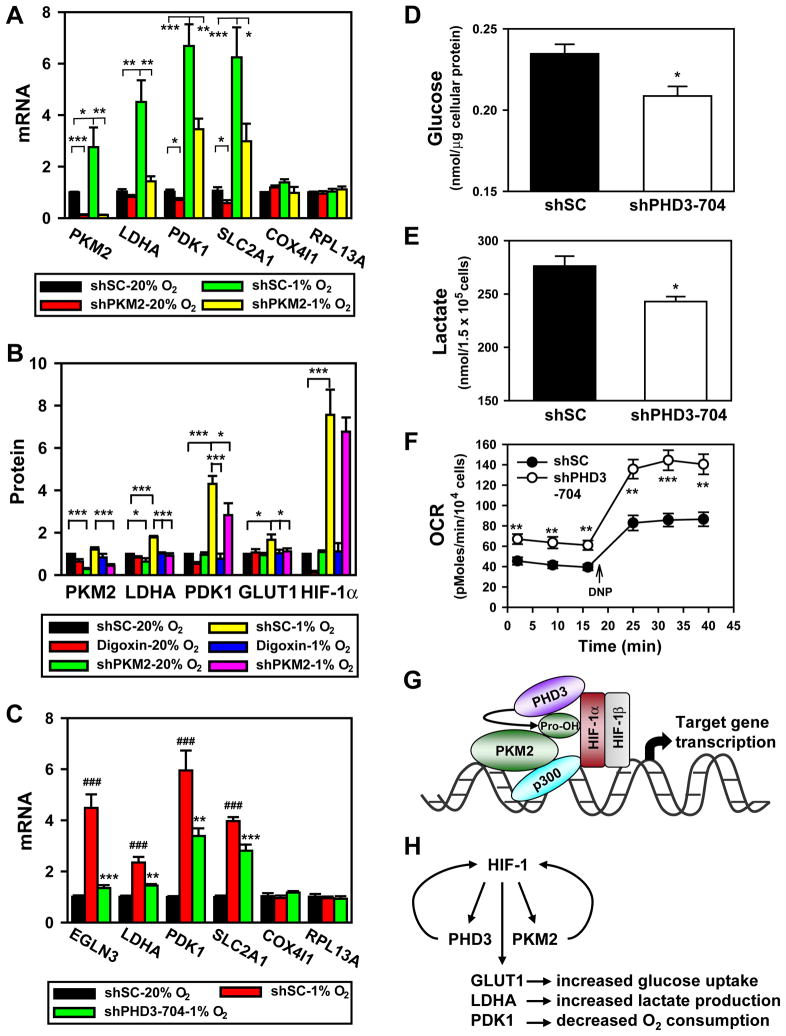
Figure 7. PKM2 and PHD3 Promote HIF-1-dependent Glycolytic Metabolism.
(A) qRT-PCR analysis of indicated mRNAs in HeLa cells transfected with shSC or shPKM2-#1 vector and exposed to 20% or 1% O2 for 24 h (mean ± SEM, n = 4). *p<0.05, **p<0.01, ***p<0.001.
(B) HeLa cells were transduced with shSC or shPKM2-#1 and shPKM2-#2 retrovirus, and exposed to 20% or 1% O2 in the absence or presence of digoxin (100 nM) for 24 h. Levels of the indicated protein were determined by immunoblot assay, quantified by densitometry, and normalized to actin (mean ± SEM, n = 4). *p<0.05, ***p<0.001.
(C) qRT-PCR analysis of indicated mRNAs in HeLa cells transduced with shSC or shPHD3-704 retrovirus and exposed to 20% or 1% O2 for 24 h (mean ± SEM, n = 3–4). ###p<0.001 vs shSC-20% O2; **p<0.01, ***p<0.001 vs shSC-1% O2.
(D–F) RCC4 cells were transduced with shSC or shPHD3-704 retrovirus. Glucose was measured in lysates and normalized to total cellular protein amount (mean ± SEM, n = 3; D); lactate was measured in the culture media and normalized to cell number (mean ± SEM, n = 3; E); O2 consumption rate (OCR) was measured and normalized to cell number (mean ± SEM, n = 4–8; F). *p<0.05, **p<0.01, ***p<0.01 vs shSC.
(G) PKM2 is prolyl hydroxylated by PHD3 and interacts with HIF-1α and p300, thereby enhancing HRE occupancy by HIF-1 and p300, and increasing histone acetylation.
(H) Feed-forward mechanism for HIF-1 activity. HIF-1 activates transcription of genes encoding PHD3 and PKM2, which interact with HIF-1α to stimulate transactivation of target genes encoding GLUT1, LDHA, PDK1, and other metabolic enzymes that mediate the Warburg effect in cancer cells.
See also Figure S6.