Abstract
Background
The analysis of human Y-chromosome variation in the context of population genetics and forensics requires the genotyping of dozens to hundreds of selected single-nucleotide polymorphisms (SNPs). In the present study, we developed a 121-plex (121 SNPs in a single array) TaqMan array capable of distinguishing most haplogroups and subhaplogroups on the Y-chromosome human phylogeny in Europe.
Results
We present data from 264 samples from several European areas and ethnic groups. The array developed in this study shows >99% accuracy of assignation to the Y human phylogeny (with an average call rate of genotypes >96%).
Conclusions
We have created and evaluated a robust and accurate Y-chromosome multiplex which minimises the possible errors due to mixup when typing the same sample in several independent reactions.
Background
The development of high-throughput technologies to genotype hundreds of thousands of markers has yielded an increase in the understanding of the genetic diversity of our species [1]. This genomic knowledge has been applied to different fields, from biomedical and pharmaceutical research to population genetics and forensics [2-5]. Most of the genotyping technologies have been based on typing of single-nucleotide polymorphisms (SNPs) that commonly have only two alleles (ancestral or derived compared to nonhuman primates) and have usually arisen once. These high-throughput SNP genotyping analyses provide much information about the variation of our genome, but still little information has been derived from some specific genome regions of great interest. SNPs are also used for human identification purposes and to reconstruct human demographic history, although these fields require the typing of a few selected SNPs for targeted research rather than the use of high-throughput genotyping methods (that is, allele-specific probes or single base primer extensions).
The human Y chromosome has been extensively analysed in forensic and evolutionary studies because of its unique properties. Despite being a complex chromosome with highly repetitive sequences [6], its exclusively paternal inheritance due to the lack of recombination over most of the chromosome has allowed researchers to trace paternal lineages and reconstruct the male demographic history of populations [7]. The human Y chromosome contains hundreds of well-characterised SNPs (around 600) whose evolutionary relationships have been established in a robust phylogeny [8]. The Y-chromosome phylogeny defines several main branches (haplogroups) named with alphabetical (A to T) and numerical codes. Each branch is subdivided into more minor braches (subhaplogroups) defined by other SNPs in a hierarchical way [9]. The classification of a male DNA sample within the Y-chromosome phylogeny requires the successive typing of markers that define the haplogroup and subhaplogroups to which the sample belongs. This process is not only laborious but also time-consuming. For this reason, several attempts to genotype multiple Y-chromosome SNPs in a single reaction have been reported, mainly using single-base extension methodologies [10-15] or oligonucleotide ligation assays [16]. Nonetheless, these multiplex approaches have not been able to type successfully more than 35 Y-chromosome SNPs in a single reaction.
It is preferable to genotype, in a reproducible manner, around 100 SNPs at a time. We therefore aimed to design a single multiplex reaction of this size using TaqMan probes (Applied Biosystems, Inc., Foster City, CA, USA). TaqMan probes are robust and reproducible assays that have been used extensively to type SNPs in single reactions, that is, one SNP per TaqMan reaction. Each assay is based on TaqMan probes that are hydrolysis probes that anneal within a specific DNA region amplified by polymerase chain reactions (PCRs). The probes present a fluorophore (usually VIC or 6-FAM, Applied Biosystems) attached to the 5′-end and a quencher at the 3′-end that prevents fluorescence of the fluorophore. As the AmpliTaq Gold DNA Polymerase (Applied Biosystems) extends the primer, its exonuclease activity degrades the probe that has annealed to the template and releases the fluorophore, relieving the quenching effect and allowing fluorescence of the fluorophore. In a TaqMan assay, two probes labelled with different fluorophores are used, each one complementary to one of the two alleles (ancestral or derived) of a SNP. In this way, in a real-time PCR, the alleles of one SNP of a DNA sample can be interrogated using two TaqMan probes labelled with different fluorophores. In the present study, our goal was to perform more than 100 TaqMan assays in a single array to define detailed haplogroups and subhaplogroups of the human Y chromosome in European populations.
Results
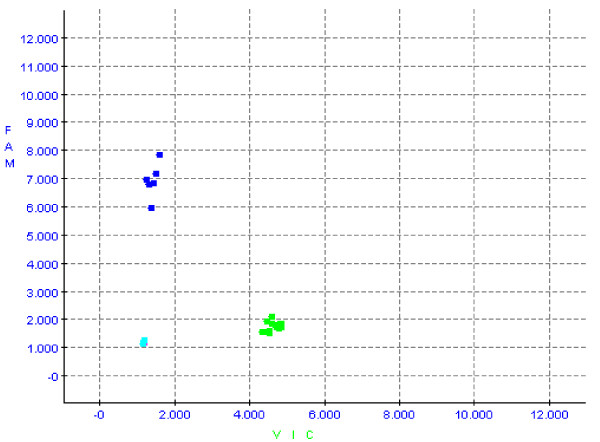
Of the 128 SNPs designed in the TaqMan OpenArray plate (Applied Biosystems, Inc.), 121 were successfully typed. The internal controls gave completely concordant results, as M145 genotypes were always concordant with M203, and the two different assays for the marker M9 gave identical results in all the individuals. In addition, the genotypes obtained for the control samples (Coriell samples; Coriell Institute for Medical Research, Camden, NJ 08103 USA) were identical to those previously obtained with the same TaqMan assays used one-by-one, giving a concordance rate of 100% (Table 1). An example of a successful assay for the M203 marker is shown in Figure 1. Three clusters of samples are shown in the plot from the Autocaller software (Applied Biosystems): those with VIC fluorescence (representing the ancestral allele, G, of the M203 marker); those with FAM fluorescence (representing the derived allele, C); and the negative controls, nontemplate controls (NTCs) without VIC or FAM fluorescence.
Table 1.
Genetic markers analysed in the OpenArray, the Y haplogroup they define, the alleles and their call rates (successful genotype calls)
| List of genetic markers analysed | List of genetic markers analysed (continuation) | List of genetic markers analysed (continuation) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | Haplogroupa | Allelesb | Call rate (%) | Marker | Haplogroupa | Allelesb | Call rate (%) | Marker | Haplogroupa | Allelesb | Call rate (%) |
| M216 | C | C/T | 95.8 | P259 | I1d | T/G | 96.6 | M231 | N | G/A | 94.7 |
| M168 | CT | C/T | 98.1 | P215 | I2 | A/G | 95.1 | M128 | N1a | 2-bp del | 97.0 |
| M174 | D | T/C | 97.7 | P37.2 | I2a | T/C | 96.6 | P43 | N1b | G/A | 96.6 |
| M145c | DE | G/A | 96.6 | P41.2 | I2a1 | T/C | 97.3 | P105 | N1c | G/A | 96.2 |
| M203c | DE | G/C | 96.6 | M26 | I2a2 | G/A | 95.5 | M214 | NO | G/C | 96.6 |
| M96 | E | G/C | 98.1 | M223 | I2b | C/T | 96.2 | M119 | O1a | A/C | 99.2 |
| P147 | E1 | T/A | 97.7 | M284 | I2b1 | ACAA/del | 96.2 | P31 | O2 | T/C | 95.8 |
| M33 | E1a | A/C | 95.5 | M379 | I2b2 | GT/del | 97.0 | M122 | O3 | T/C | 98.9 |
| P177 | E1b | C/T | 93.6 | P95 | I2b4 | G/T | 95.1 | M45 | P | G/A | 96.2 |
| P2 | E1b1 | C/T | 97.3 | P123 | IJ | T/C | 94.3 | M242 | Q | C/T | 97.3 |
| M2 | E1b1a | A/G | 97.0 | M304 | J | A/C | 94.3 | M207 | R | A/G | 95.5 |
| M215 | E1b1b | A/G | 97.3 | M267 | J1 | T/G | 95.5 | M173 | R1 | A/C | 93.2 |
| M35 | E1b1b1 | G/C | 95.8 | M62 | J1a | T/C | 95.5 | SRY10831.2 | R1a | G/A | 99.6 |
| M78 | E1b1b1a | C/T | 98.1 | M365 | J1b | A/G | Failed | M56 | R1a1a | A/T | 97.7 |
| V12 | E1b1b1a1 | A/G | 97.3 | M390 | J1c | A/ins | Failed | M157 | R1a1b | A/C | 98.1 |
| V13 | E1b1b1a2 | G/A | 97.0 | P56 | J1d | A/G | 96.6 | M204 | R1a1c | T/G | 95.8 |
| V22 | E1b1b1a3 | T/C | 97.7 | P58 | J1e | T/C | 95.5 | P98 | R1a1d | C/T | Failed |
| V65 | E1b1b1a4 | G/T | 96.2 | M172 | J2 | T/G | 96.6 | PK5 | R1a1e | C/T | 95.8 |
| M81 | E1b1b1b | C/T | 89.8 | M410 | J2a | A/G | 95.5 | M343 | R1b | C/A | 97.3 |
| M123 | E1b1b1c | G/A | 97.7 | M47 | J2a1 | G/A | 96.6 | M18 | R1b1a | 2-bp ins | 96.6 |
| M281 | E1b1b1d | G/A | 97.0 | M340 | J2a10 | G/C | 94.7 | P297 | R1b1b | G/C | 95.8 |
| V6 | E1b1b1e | G/C | 99.6 | M419 | J2a11 | AAAAG/del | 95.8 | M73 | R1b1b1 | 2-bp del | 96.2 |
| P72 | E1b1b1f | G/A | 98.1 | P81 | J2a12 | C/T | Failed | M269 | R1b1b2 | C/T | 96.2 |
| M329 | E1b1c | G/C | 98.1 | P279 | J2a13 | G/A | 98.1 | L23 | R1b1b2a | A/Ge | 84.8 |
| P75 | E1b2 | G/A | Failed | M67 | J2a2 | A/T | 95.1 | P311 | R1b1b2a1 | A/G | 97.0 |
| M75 | E2 | G/A | 97.7 | M92 | J2a2a | T/C | 96.2 | U106 | R1b1b2a1a | C/T | 97.3 |
| M89 | F | C/T | 97.7 | M68 | J2a3 | A/G | 91.3 | U198 | R1b1b2a1a1 | G/A | 98.5 |
| M201 | G | G/T | 95.5 | M137 | J2a4 | T/C | 97.7 | P107 | R1b1b2a1a2 | G/A | Failed |
| M285 | G1 | G/C | 96.6 | M158 | J2a5 | G/A | 97.7 | P312 | R1b1b2a1b | A/Ce | 97.3 |
| P20 | G1a | C/del | 95.8 | M289 | J2a6 | G/A | 96.6 | M65 | R1b1b2a1b1 | A/T | 96.6 |
| P76 | G1b | G/C | 98.9 | M318 | J2a7 | T/C | 97.3 | M153 | R1b1b2a1b2 | T/A | 97.7 |
| P287 | G2 | G/T | 96.6 | M319 | J2a8 | T/A | 97.7 | SRY2627 | R1b1b2a1b3 | C/T | 97.3 |
| P15 | G2a | C/T | 97.7 | M339 | J2a9 | T/G | 97.7 | U152 | R1b1b2a1b4 | C/T | 97.0 |
| M287 | G2b | A/T | 97.0 | M12 | J2b | G/T | 97.0 | M126 | R1b1b2a1b4a | 4-bp del | 97.3 |
| M377 | G2c | A/G | 94.7 | M205 | J2b1 | T/A | 98.1 | M160 | R1b1b2a1b4b | A/C | 96.2 |
| M69 | H | T/C | 97.0 | M241 | J2b2 | G/A | Failed | L21 | R1b1b2a1b5 | C/G | 97.7 |
| M52 | H1 | A/C | 95.5 | M9ad | K | C/G | 97.3 | M37 | R1b1b2a1b5a | C/T | 87.9 |
| Apt | H2 | G/A | 92.8 | M9bd | K | C/G | 97.3 | M222 | R1b1b2a1b5b | G/A | 97.7 |
| M170 | I | A/C | 97.0 | M20 | L | A/G | 94.3 | P66 | R1b1b2a1b5c | G/A | 98.1 |
| M253 | I1 | C/T | 97.0 | M27 | L1 | C/G | 94.7 | M335 | R1b1c | T/A | 96.2 |
| M21 | I1a | A/T | 96.2 | M317 | L2 | GA/del | 94.3 | M124 | R2 | C/T | 98.1 |
| M72 | I1b1 | A/G | 98.1 | M357 | L3 | C/A | 97.3 | M70 | T | A/C | 97.0 |
| P109 | I1c | G/A | 95.8 | P256 | M | G/A | 97.7 | ||||
aHaplogroups are based upon data published by Karafet et al. [8] and on the Internal Society of Genetic Genealogy (ISOGG) 2009 Y-DNA SNP Index website (http://isogg.org/tree/ISOGG_YDNA_SNP_Index09.html).
bThe two alleles of each marker (ancestral or derived) are shown.
cBoth markers (M145 and M203) define the same branch of the human Y-chromosome phylogeny (haplogroup DE).
dMarker M9 was genotyped with two independent assays, with TaqMan probes labelled with different fluorophores (see "SNP selection" inside the Material and Methods section).
eAncestral allele not available.
Figure 1.
Autocaller plot showing the fluorescence of 22 samples (green and dark blue) and two nontemplate controls (light blue) in the M203 assay. The TaqMan probe complementary to the ancestral allele (G) was labelled with the VIC fluorophore, whereas FAM was used to label the derived allele (C). Three clusters are seen in the plot: the samples with the derived allele, thus belonging to the Y-chromosome haplogroup branch DE, are shown in dark blue; the samples with the ancestral allele, thus belonging to any haplogroup except DE, are shown in green; and the negative controls are shown in light blue.
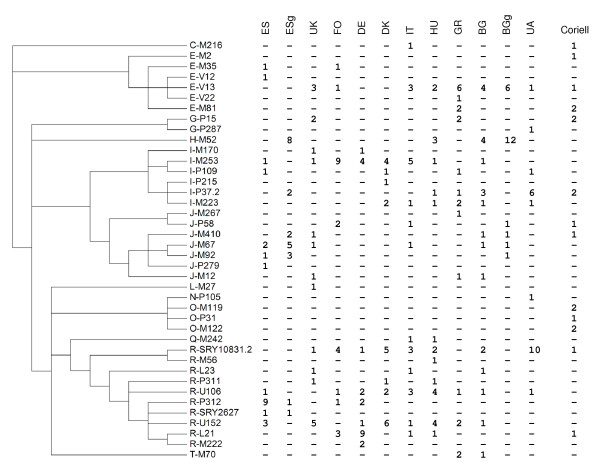
The number of successful genotypes for the remaining 121 SNPs in the pool (the call rate) was high (average 96.4%), with the lowest value being for L23 (84.8%) (Table 1). The combination of 121 SNPs was able to distinguish a total of 118 different haplogroups and subhaplogroups (Additional file 1, Figure S1), 40 of which were present in the populations typed and 24 of which were shared by two or more European samples (Figure 2). Haplogroup composition varied greatly across the different populations sampled, as expected from the known phylogeography of the samples used.
Figure 2.
Phylogenetic tree showing the haplogroup counts found in the present sample set following the nomenclature published by Karafet et al. [8] and on the Internal Society of Genetic Genealogy (ISOGG) 2009 Y-DNA SNP Index website (http://isogg.org/tree/ISOGG_YDNA_SNP_Index09.html). ES, Spanish; ESg, Spanish Roma; UK, British; FO, Faroe Islands; DE, German; DK, Danish; IT, Italian; HU, Hungarian; GR, Greek; BG, Bulgarian; BGg, Bulgarian Roma; UA, Ukrainian.
Of 282 individuals where genotyping was attempted, three did not yield any amplified product for any assay. This failure was due to technical manipulation problems in the DNA loading phase, as the wells corresponding to these three samples were empty in the array. Of the remaining 279, 277 showed complete phylogenetic compatibility. If a sample showed the derived allele for a SNP that led to a certain branch of the tree, it also presented the derived alleles for all the SNPs leading to that branch and the ancestral alleles for the SNPs leading to other branches. For example, if a sample was derived for M201 (allele T), it was also derived for M89 (allele T), M168 (allele T), M42 (allele T) and SRY10831.1 (allele G), but was ancestral for all the rest of the SNPs in the array (see Table 1). Although some of the haplogroups (that is, haplogroups D and M) were not present in the populations sampled, all the samples typed showed the ancestral allele for the SNPs defining these branches. Two individuals showed phylogenetic incompatibilities; that is, we found derived alleles for SNPs leading to different branches of the phylogeny in the same individual. In both cases, this incongruence was due to a single misassigned internal SNP (false-positive) that was eliminated, and both individuals could be correctly assigned in the phylogeny. However, it should be noted that if the misassignation occurs at the very end of the branch where the sample is assigned in the phylogeny, it might be undetectable and would lead to the wrong assignation at the subbranch level.
Discussion
The aim of the present study was to create a SNP multiplex for the human Y chromosome in a single array that could be used to define Y-chromosome haplogroups in European populations. Previously, genotyping of Y-chromosome SNPs has been performed in single-plex reactions or using limited multiplexes [10-16]. The multiplex SNP typing described in the present study greatly reduces the amount of time spent on typing. Besides that, typing a large number of SNPs in a single array avoids the possibility of errors by sample mixup when typing the same sample in several independent reactions. The pass rate (>94% successfully genotyped SNPs) and the average call rate (>96% successful genotypes) in our study are remarkable and confirm the present multiplex SNP typing as a robust method. The analysis of the human Y chromosome has some technical advantages compared to other genomic regions, which could explain the high pass and call rates. Despite the complex and highly repetitive structure of the human Y chromosome [6], only one allele for each marker per individual is expected because of its uniparental inheritance and the choice of markers originating from unique regions. All male samples are thus hemizygous (presenting only one of the two alleles), and no heterozygous results are expected (or observed). This simplifies the calling, since only two clusters of results are expected (excluding the cluster of negative controls), one for each allele. Other genomic regions might be more difficult to genotype with the present technique, since the call rate must distinguish homozygous from heterozygous alleles.
The amount of DNA used in the present sample set as recommended by the manufacturer is high (300 ng); however, high pass rates and calling rates (similar to the ones provided in the present sample set) have been obtained with as little as 90 ng (thus a final amount of 45 ng used for genotyping), provided that the A260/A280 DNA absorbance ratio is approximately 1.8 (data not shown), which stresses the relevance of the DNA purity to perform the analyses. This DNA amount is within the range described in previous works multiplexing the Y chromosome, mitochondrial DNA and autosomal SNPs [10,12,16], although it is higher than that in other studies [11,13,14]. Nonetheless, this amount of DNA remains lower than the amount needed for genome-wide DNA chips (for example, 500 ng for the Genome-Wide Human SNP Array 6.0; Affymetrix, Inc., Santa Clara, CA 95051 USA; and 37.5 μg for the next-generation Omni microarrays derived from genome-wide association studies; Illumina, Inc. (San Diego, CA 92121-1975 USA), although these cover a much larger number of markers. In addition, the amount of DNA used in this study should not be a problem for population genetic studies, as the mean amount of human DNA extracted from 1 mL of saliva has been reported to be as much as 11.4 μg/mL [20] and even larger amounts are extracted from blood. The present assay might not be useful for forensic studies with very low amounts of DNA available (for example, extraction from a single hair or from degraded samples), although it can certainly be useful for forensic cases without a limited quantity of DNA (for example, paternity and family reunification analyses).
Although the present multiplex was designed to screen populations of European origin, it might be used to detect the presence of individuals of non-European ancestry in European samples and as a first approach for other admixed populations with subsequent typing for those haplogroups not common in Europe, since some branches of the Y-chromosome phylogeny are not defined in the present assay (that is, branches A or B or subbranches within C, D, K, M, N, O, P, Q or T). Nonetheless, the present results show that the TaqMan assay multiplex technology exhibits successful and robust results and that other combination of SNPs could be designed to genotype all branches of the Y-chromosome phylogeny or to focus on specific regions of the phylogeny. The OpenArray technology has the potential to combine up to 256 TaqMan assays in the same plate, which is enough to genotype the main diagnostic SNPs of most branches and subbranches of the human Y-chromosome phylogeny. In addition, the present methodology is flexible enough to include those markers on the Y chromosome that are required for a specific analysis which contrast with the rigidity of commercial high-throughput arrays that also include several SNPs in the Y chromosome. Although several genome-wide human SNP arrays have incorporated hundreds of Y-chromosome SNPs, just a few of the SNPs in our array share commonality with some commercially available SNP arrays (for instance, M216, M9 and P123 are included in the Genome-Wide Human SNP Array 6.0 manufactured by Affymetrix, Inc.; and M145 and M173 are included in the BeadArray Reader manufactured by Illumina, Inc.). Thus, to our knowledge, this study is the first to describe typing with flexibility to such a high accuracy the human Y chromosome in a single array.
Conclusions
We have created and evaluated a robust and accurate Y-chromosome array to genotype 121 SNPs at a time using TaqMan probes that classifies individuals into the main haplogroups and the main European subhaplogroups in the human Y-chromosome phylogeny, substantially decreasing time of laboratory work and minimising the possible errors due to mixup when typing the same sample in several independent reactions.
Methods
Samples
DNA samples were obtained from 22 healthy unrelated males from each of the following 12 populations: Bulgarians, Bulgarian Roma, Spanish, Spanish Roma, Italians, Germans, British, Faroe Islanders, Danish, Greeks, Hungarians and Ukrainians. DNA samples from another 18 individuals of diverse origins obtained from the Coriell Institute for Medical Research (three from Taiwan, one Mbuti Pygmy, seven Russians, two Basques, one Chinese, two from the Middle East and two from Southeast Asia) were used as internal controls of the accuracy of the multiplex, since their haplogroup affiliations had previously been determined by two different laboratories using the same SNPs in single reactions incorporated into the present multiplex assay. Informed consent was obtained from all participants, and the project was approved by the Clinical Research Ethics Committee at the Institut Municipal d'Assistència Sanitària (IMAS reference 2006/2600/I). DNA extraction was carried out using a standard phenol/chloroform method.
SNP selection
A total of 128 Y-chromosome SNPs defining the main haplogroups and subhaplogroups were chosen, with special attention to SNPs defining haplogroups and subhaplogroups reported as variable in European samples [17-19]. Data published by Karafet et al. [8] and four SNPs (P311, P312, L21, and L23) published on the Internal Society of Genetic Genealogy (ISOGG) 2009 Y-DNA SNP Index website (http://isogg.org/tree/ISOGG_YDNA_SNP_Index09.html) were used for SNP selection. The flanking sequences for each of the SNPs were then investigated for the design of the SNP TaqMan probes and submitted to the TaqMan assay design pipeline (Applied Biosystems, Inc.). See Additional file 2, Table S2, for a complete description of the primers used. Two different control assays were included in the multiplex. Two redundant assays (M145 and M203) for haplogroup DE, that is, SNPs that define the same Y-chromosome branch, were included as an internal phylogenetic control. In addition, two different assays for the same SNP (M9) were designed as an additional internal control. An M9a assay was designed to present the ancestral allele (C) in the probe with the VIC dye and the derived allele (G) with the FAM dye. The M9b assay was designed to present the ancestral allele (C) in the probe with the FAM dye and the derived allele (G) with the VIC dye.
SNP genotyping and haplogroup classification
In the present analysis, a multiplex of 128 TaqMan assays was performed in a single array for each DNA sample. Each TaqMan OpenArray plate was designed to contain the 128 assays for a total of 24 samples (22 DNA samples and 2 NTCs). Technical exigencies of the autoloader machine require double the amount of DNA to be present in the loading tips. A total of 300 ng of genomic DNA was used, and a final amount of 150 ng was incorporated into the array with the autoloader and genotyped according to the manufacturer's recommendations. The multiplex TaqMan assay reactions were carried out in a dual 384-well GeneAmp 9700 Thermal Cycler (Applied Biosystems, Inc.) with the following PCR cycle: an initial step at 93°C for 10 minutes followed by 55 cycles of 45 seconds at 95°C, 13 seconds at 94°C and 2 minutes, 14 seconds at 53°C.
The fluorescence results were read using Autocaller software (Applied Biosystems, Inc.). The genotypes were compiled and used to assign each sample to its Y-chromosome haplogroup according to the Y-chromosome phylogeny published by Karafet et al. [8] and the Internal Society of Genetic Genealogy (ISOGG) 2009 Y-DNA SNP Index website (http://isogg.org/tree/ISOGG_YDNA_SNP_Index09.html).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BMC and DC planned the study and wrote the manuscript. BMC analysed the data. JZ designed the assays. PS, GS, RA and SP performed the laboratory work. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Phylogenetic tree showing the haplogroup tested with the OpenArray following the nomenclature published by Karafet et al. [8] and on the Internal Society of Genetic Genealogy (ISOGG) 2009 Y-DNA SNP Index website (http://isogg.org/tree/ISOGG_YDNA_SNP_Index09.html). Recurrent mutations of three markers (SRY10831, P37 and P41) are shown in different branches of the tree.
Table S2. List of the primers used for the amplification of every assay implemented in the OpenArray.
Contributor Information
Begoña Martínez-Cruz, Email: begonya.martinez@upf.edu.
Janet Ziegle, Email: Janet.Ziegle@lifetech.com.
Paula Sanz, Email: paula.sanz@upf.edu.
Graciela Sotelo, Email: graciela.sotelo@upf.edu.
Roger Anglada, Email: roger.anglada@upf.edu.
Stéphanie Plaza, Email: stephanie.plaza@upf.edu.
David Comas, Email: david.comas@upf.edu.
Acknowledgements
We thank Mònica Vallés (UPF) for her technical help and Doron Behar (Molecular Medicine Laboratory, Rambam Health Care Campus, Haifa, Israel) for sharing unpublished data. We are indebted to everyone who contributed samples to this study: Antonella Useli, Donata Luiselli and Davide Petener (Università di Bologna); Anatasia Kouvatsi (Aristotle University of Thessaloniki); Halyna Makukh (Institute of Hereditary Pathology of the Ukrainian Academy of Medical Sciences, Lviv, Ukraine); Horolma Pamjav (Institut Forensic Medicine, Budapest, Hungary); Dora Angelicheva and Luba Kalaydjieva (The University of Western Australia, Perth, Australia); David M Hougaard, Mads V Hollegaard and Bent Norgaard-Pedersen (Statens Serum Institut, Copenhagen, Denmark); Hanne Strager (Natural History Museum of Denmark, Copenhagen, Denmark); Henning Bundgaard (National University Hospital, Copenhagen, Denmark); Eske Willerslev (University of Copenhagen); and the Spanish Banco Nacional de ADN (Centre of Cancer Research, University of Salamanca). The present study was supported by the Ministerio de Ciencia e Innovación (Madrid, Spain) (CGL2010-14944) and Direcció General de Recerca, Generalitat de Catalunya (Barcelona, Spain) (2009SGR1101).
The Genographic Consortium members: Syama Adhikarla1, Christina J Adler2, Danielle A Badro3, Elena Balanovska4, Oleg Balanovsky4, Jaume Bertranpetit5, Andrew C Clarke6, Alan Cooper2, Clio SI Der Sarkissian2, Matthew C Dulik7, Christoff J Erasmus8, Jill B Gaieski7, ArunKumar GaneshPrasad1, Wolfgang Haak2, Marc Haber3, Angela Hobbs8, Asif Javed9, Li Jin10, Matthew E Kaplan11, Shilin Li10, Elizabeth A Matisoo-Smith6, Marta Melé5, Nirav C Merchant11, R John Mitchell12, Amanda C Owings7, Laxmi Parida9, Ramasamy Pitchappan1, Daniel E Platt9, Lluis Quintana-Murci13, Colin Renfrew14, Daniela R Lacerda15, Ajay K Royyuru9, Fabrício R Santos15, Theodore G Schurr7, Himla Soodyall8, David F Soria Hernanz16, Pandikumar Swamikrishnan17, Chris Tyler-Smith18, Kavitha Valampuri John1, Arun Varatharajan Santhakumari1, Pedro Paulo Vieira19, Pierre A Zalloua3 and R Spencer Wells16.
References
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao O, Lu TT, Nothnagel M, Junge O, Freitag-Wolf S, Caliebe A, Balascakova M, Bertranpetit J, Bindoff LA, Comas D, Holmlund G, Kouvatsi A, Macek M, Mollet I, Parson W, Palo J, Ploski R, Sajantila A, Tagliabracci A, Gether U, Werge T, Rivadeneira F, Hofman A, Uitterlinden AG, Gieger C, Wichmann HE, Rüther A, Schreiber S, Becker C, Nürnberg P. et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- Adeyemo A, Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics. 2010;13:72–79. doi: 10.1159/000218711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Jobling MA. Y-chromosomal SNP haplotype diversity in forensic analysis. Forensic Sci Int. 2001;118:158–162. doi: 10.1016/S0379-0738(01)00385-1. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou SF, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krjutskov K, Viltrop T, Palta P, Metspalu E, Tamm E, Suvi S, Sak K, Merilo A, Sork H, Teek R, Nikopensius T, Kivisild T, Metspalu A. Evaluation of the 124-plex SNP typing microarray for forensic testing. Forensic Sci Int Genet. 2009;4:43–48. doi: 10.1016/j.fsigen.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Brión M, Sobrino B, Blanco-Verea A, Lareu MV, Carracedo A. Hierarchical analysis of 30 Y-chromosome SNPs in European populations. Int J Legal Med. 2005;119:10–15. doi: 10.1007/s00414-004-0439-2. [DOI] [PubMed] [Google Scholar]
- Paracchini S, Arredi B, Chalk R, Tyler-Smith C. Hierarchical high-throughput SNP genotyping of the human Y chromosome using MALDI-TOF mass spectrometry. Nucleic Acids Res. 2002;30:6. doi: 10.1093/nar/30.2.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brión M, Sanchez JJ, Balogh K, Thacker C, Blanco-Verea A, Børsting C, Stradmann-Bellinghausen B, Bogus M, Syndercombe-Court D, Schneider PM, Carracedo A, Morling N. Introduction of a single nucleotide polymorphism-based "Major Y-chromosome haplogroup typing kit" suitable for predicting the geographical origin of male lineages. Electrophoresis. 2005;26:4411–4420. doi: 10.1002/elps.200500293. [DOI] [PubMed] [Google Scholar]
- Sanchez JJ, Børsting C, Hallenberg C, Buchard A, Hernandez A, Morling N. Multiplex PCR and minisequencing of SNPs: a model with 35 Y chromosome SNPs. Forensic Sci Int. 2003;137:74–84. doi: 10.1016/S0379-0738(03)00299-8. [DOI] [PubMed] [Google Scholar]
- Lessig R, Zoledziewska M, Fahr K, Edelmann J, Kostrzewa A, Dobosz T, Kleemann WJ. Y-SNP-genotyping: a new approach in forensic analysis. Forensic Sci Int. 2005;154:128–136. doi: 10.1016/j.forsciint.2004.09.129. [DOI] [PubMed] [Google Scholar]
- Berniell-Lee G, Sandoval K, Mendizabal I, Bosch E, Comas D. SNPlexing the human Y-chromosome: a single-assay system for major haplogroup screening. Electrophoresis. 2007;28:3201–3206. doi: 10.1002/elps.200700078. [DOI] [PubMed] [Google Scholar]
- Francalacci P, Sanna D. History and geography of human Y-chromosome in Europe: a SNP perspective. J Anthropol Sci. 2008;86:59–89. [PubMed] [Google Scholar]
- Francalacci P, Morelli L, Useli A, Sanna D. The history and geography of the Y chromosome SNPs in Europe: an update. J Anthropol Sci. 2010;88:207–214. [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Lahr MM, Foley RA, Oefner PJ, Cavalli-Sforza LL. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- Quinque D, Kittler R, Kayser M, Stoneking M, Nasidze I. Evaluation of saliva as a source of human DNA for population and association studies. Anal Biochem. 2006;353:272–277. doi: 10.1016/j.ab.2006.03.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic tree showing the haplogroup tested with the OpenArray following the nomenclature published by Karafet et al. [8] and on the Internal Society of Genetic Genealogy (ISOGG) 2009 Y-DNA SNP Index website (http://isogg.org/tree/ISOGG_YDNA_SNP_Index09.html). Recurrent mutations of three markers (SRY10831, P37 and P41) are shown in different branches of the tree.
Table S2. List of the primers used for the amplification of every assay implemented in the OpenArray.