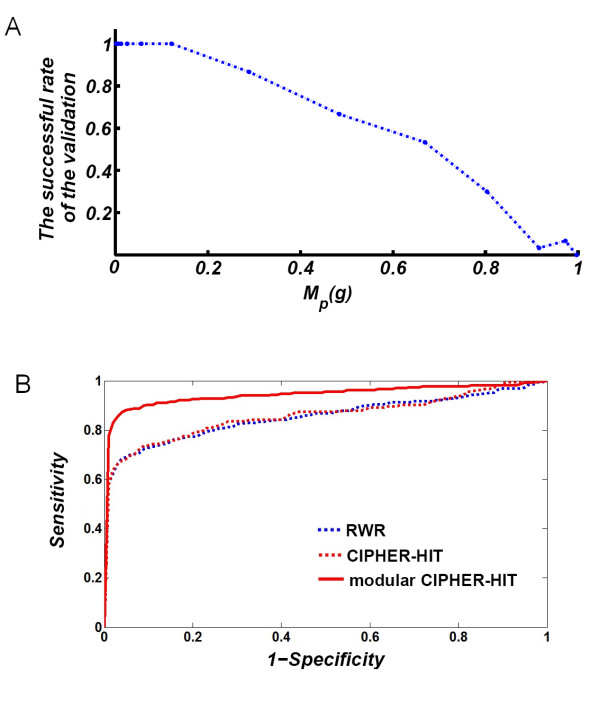
Figure 3.
Results of the genome-wide cross-validation for disease gene prioritization. (A) The conditional Mean-Hitting-Time (Mp(g)) is calculated by Equation (5). Results showed that genes with high modularity levels to the other adjacent nodes with small Mp(g) values will be more likely to be successfully prioritized during the validation. (B) The receiver operating characteristic (ROC) curves of the genome-wide leave-one-out cross-validation. The horizontal coordinates (1-Specificity) refer to values of θR, while the vertical coordinates (Sensitivity) refer to the true-positive rate corresponding to θR. The red solid line denotes the inference of modular CIPHER-HIT based on the nodes in 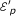 , i.e. only the adjacent node u of Mp(u) <θM = 0.3 are used for inference. The dashed lines both denote the inference based on the nodes in
, i.e. only the adjacent node u of Mp(u) <θM = 0.3 are used for inference. The dashed lines both denote the inference based on the nodes in 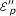 , i.e. only the adjacent node u of Mp(u) ≥ θM = 0.3 are used for inference, where the blue dashed line denotes results from the random walk with restarts (RWR) method, and the red dashed line denotes results from the CIPHER-HIT method.
, i.e. only the adjacent node u of Mp(u) ≥ θM = 0.3 are used for inference, where the blue dashed line denotes results from the random walk with restarts (RWR) method, and the red dashed line denotes results from the CIPHER-HIT method.