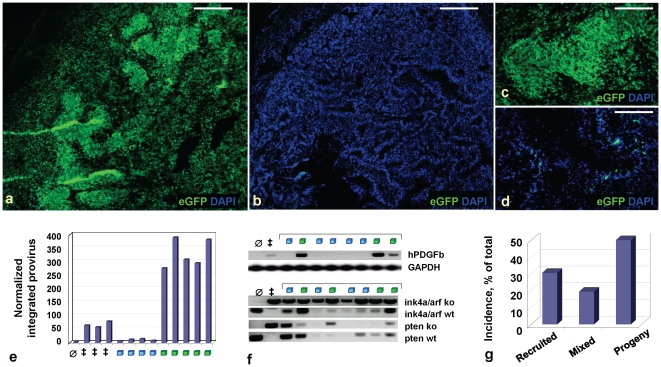
Figure 3. GBMs arising in mice targeted for heterozygous tumor suppressor loss show expansion of either recruited or progeny cell populations in pseudopalisade regions.
DAPI-stained composite (a,b) and 20X (c,d) images of frozen sections showing virally encoded eGFP in RCAS-PSG-induced Ntv-a GBMs heterozygous for loss of Ink4a, Arf and Pten at tumor initiation, with stochastic tumor suppressor loss in regions of progeny (a,c) or recruited (b,d) cells during tumor progression. (e) real-time qPCR for viral hPDGFb-HA on tumor cell DNA extracted from mouse GBMs, indicating much lesser amounts of viral integrations in GBMs containing recruited pseudopalisades (blue squares), as compared to progeny pseudopalisades (green squares). Graph shows fold change over the background in the uninjected adult mouse brain (∅); all analyses are normalized to GAPDH as a control. ‡, positive control, RCAS-PSG Ntv-a GBMs deleted for Ink4a/Arf and Pten. (f) PCR for viral hPDGFb-HA, wild-type or knockout Ink4a/Arf loci and Pten alleles on tumor cell DNA extracted from mouse glioma sections. hPDGFb-HA PCR: ∅, negative control, uninjected adult mouse brain; ‡, positive control, Ntv-a GBM homozygous deleted for Ink4a, Arf and Pten. GAPDH PCR was done in the same tube. Ink4a/Arf, Pten PCR: ∅ and ‡ indicate controls for wild-type or knockout alleles and loci, respectively (presence of band in KO lane indicates deleted allele). Gliomas that lost Ink4a/Arf during tumor progression were also likely to lose wild-type Pten allele. (g) Incidence of GBMs containing regions of recruited cells, progeny cells or a mixture of these cells in pseudopalisade regions, in Ntv-a mice heterozygous targeted for loss of Ink4a, Arf and Pten. While most GBMs arising in mice heterozygous deleted for Ink4a/Arf and Pten at glioma initiation contain large regions of progeny, about ∼30% show large areas of recruitment (total number of tumors analyzed, n = 36).