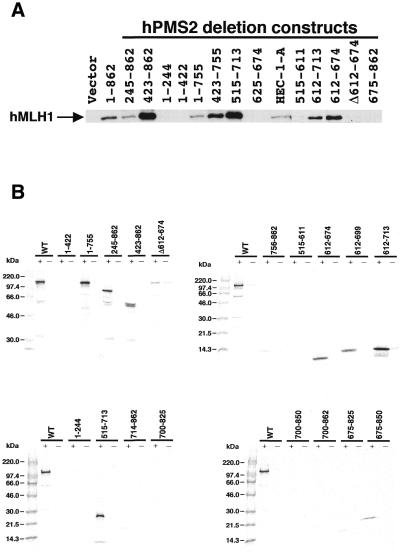
Figure 3.
Determination of the hMLH1-interactive domain of hPMS2 at residues 612–674 using the immunoprecipitation study (A) and the GST-IVTT assay (B). (A) Extracts of HEC-1-A cells transfected with pFLAG plasmids containing various hPMS2 deletion constructs were subjected to immunoprecipitation analyses using the anti-FLAG M2 monoclonal antibody. Immunoprecipitated proteins were analysed by the use of anti-hMLH1 monoclonal antibody. Lane HEC-1-A illustrates 20 µg of the cell extract that was subjected to immunoblotting without prior immunoprecipitation as the control. The arrow indicates the position of the hMLH1 protein. The minimal interaction domain of hPMS2 with hMLH1 lies between residues 612 and 674. (B) 35S-labeled full-length and deletion mutant proteins of IVTT-hPMS2 were added to glutathione beads that had been pre-treated with either GST alone (–) or GST-hMLH1 (+). Samples were resolved on 8% (top left), 15% (top right) or 10–20% (bottom) SDS-PAGE and examined by BAS-1500 and LAS-1000 (Fuji Film). Again the minimal interacting domain of hPMS2 with hMLH1 resides at residues 612–674.