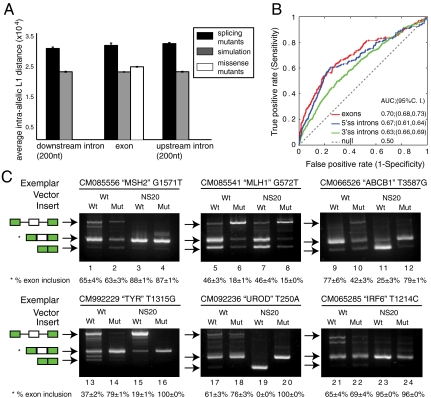
Fig. 4.
Human disease alleles are predicted to disrupt splicing. (A) Average intraallelic L1 distances for each category of mutation (HGMD splicing and HGMD missense/nonsense) and their corresponding background models of simulated mutations divided by location with respect to the splice sites. Error bars denote 95% confidence intervals. (B) Receiver operating characteristics (ROC) curve analysis using HGMD splicing mutants in regions around the 3′ss and 5′ss as “true positives” and simulated mutations as “true negatives.” ROC curve analysis classifies these mutations at decreasing thresholds of L1 stringency plotting the false against true positive rates. The exonic region is shown in red; upstream and downstream intronic regions are shown in green and blue, respectively. (C) Exemplars were selected from the HGMD missense mutants with the highest intraallelic L1 distance. Total RNA from transfection into 293 cells was analyzed by RT-PCR. The HGMD ID, gene name, and the mutational position are shown for each experiment. Quantifications on exon inclusion products are also shown. Arrows indicate the identity of the splicing product.