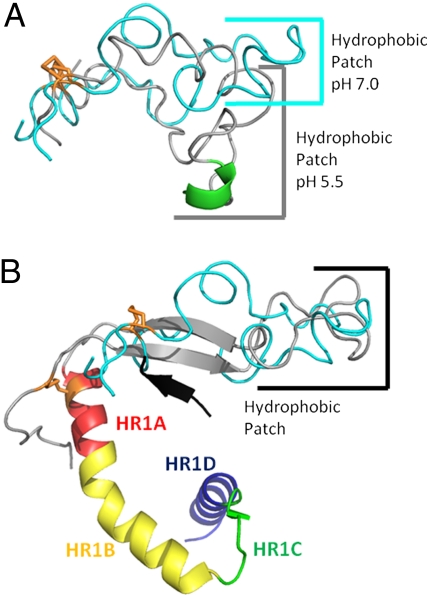
Fig. 5.
Comparisons of the Ebov FL NMR structures and a portion of the prefusion crystal structure of the trimeric GP ectodomain [Protein Data Bank (PDB) ID code 3CSY]. (A) Overlay of lowest-energy NMR conformers at pH 7.0 (cyan) and 5.5 (gray) with its helix in green. The structures are aligned by the disulfide bond shown in orange. (B) Overlay of lowest-energy NMR conformer at pH 7.0 (cyan) with Ebov fusion loop residues and HR1 from PDB ID code 3CSY (fusion loop, gray; HR1, rainbow). β-Sheet strand 6 of GP1, shown in black, presumably stabilizes the two strands of β-sheet seen in the FL of the crystal structure (corresponding to residues 515–520 and 543–548 of Fig. 1). Constructs are aligned by residues at the tip of the hydrophobic patch.