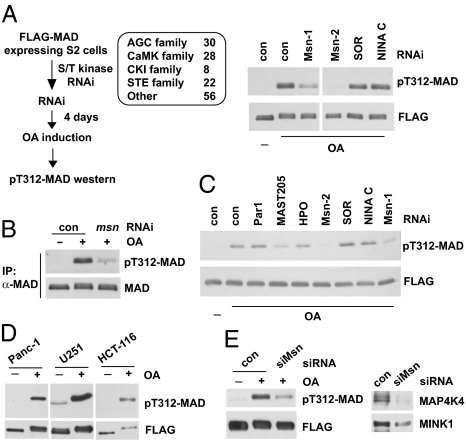
Fig. 3.
RNAi screening of the kinome uncovered Msn as the Smad α-helix 1 kinase. (A) Indicated Ser/Thr kinase families were screened as described on the left; the immunoblots show that depletion of Msn (by two independent dsRNAs, Msn-1 and Msn-2) largely diminished T312 phosphorylation of MAD in OA-treated cells. SOR and NINA C are two other kinases that scored negative in the screening. (B) After msn RNAi and OA treatment, endogenous MAD was immunoprecipitated, and T312 phosphorylation was evaluated by immunoblot analysis. TrueBlot secondary antibodies were used to avoid detecting the IgG band. (C) In vitro kinase assays showing extracts from Msn-depleted cells (Msn-1 and Msn-2 RNAi) exhibited much weaker activity in phosphorylating T312 of MAD. Par1, MAST205, and HPO are other kinases that were tested in parallel. (D) T322 phosphorylation of Smad1 in several cancer cell lines. FLAG-Smad1 was immunoprecipitated after OA treatment to measure T322 phosphorylation. The pT312-MAD antibody recognizes T322 phosphorylation of Smad1 given the shared epitopes. (E) U251 cells expressing FLAG-Smad1 was transfected with control or siRNA that depleted both MAP4K4 and MINK1 (siMsn, Right). After OA treatment, FLAG-Smad1 was immunoprecipitated, and T322 phosphorylation was examined (Left).