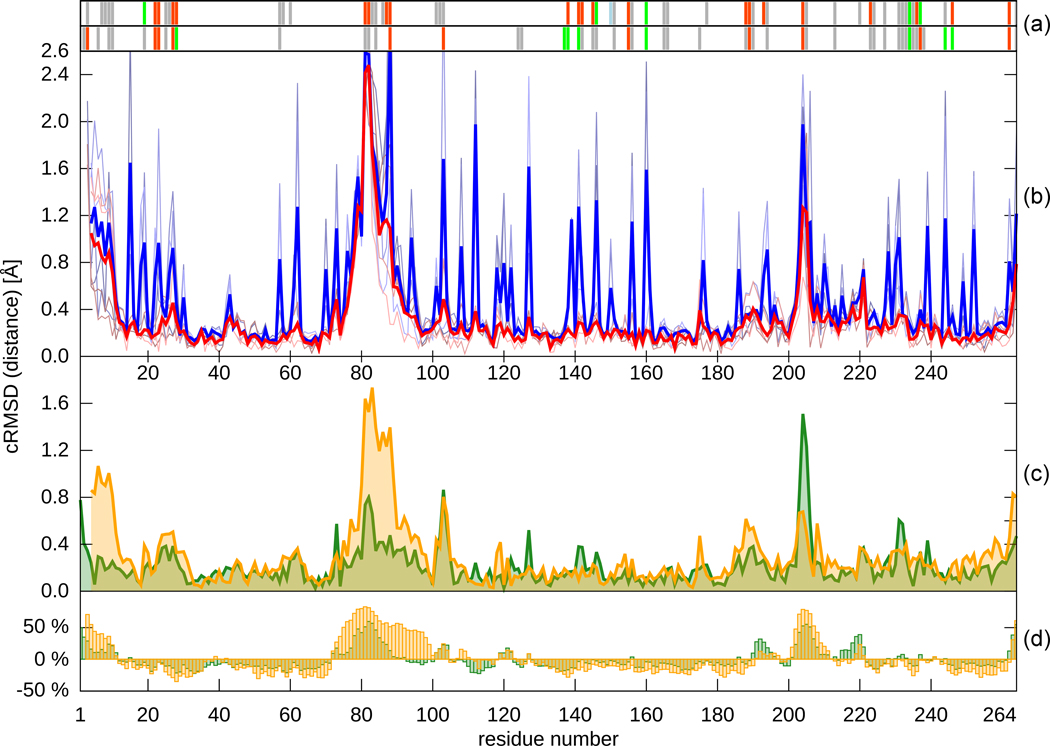
Fig. 2.
Different properties of residues in the Ba2390 structures plotted as a function of residue number. (a) A plot of the atom contacts between molecules in crystal lattice: red - residues forming close contacts (from 2.0 to 3.5 Å), green - residues forming hydrogen bonds, pale blue – residues bridged by water molecule, grey - residues forming other contacts (from 3.5 to 4.5 Å). The top graph represents contacts in the structure of the apo-form of BA2930, and the bottom in the CoA-bound structure. Contacts and distances were calculated using the CONTACT program from the CCP4 suite. (b) Plot of the RMSD and distance profiles comparing the apo and CoA-bound structures. Blue lines - RMSD values between corresponding residues. Red lines - distances between corresponding Cα atoms. The pale, thin lines represent pairwise chain comparisons between structures. Averaged results using a window of 4 comparisons are plotted with bold lines. (c) Cα distances between superposed A and B chains from one structure. The structure of the apo-form of BA2930 is represented in orange and the CoA-bound structure in green. (d) Percent deviation of the Cα temperature factors from the average value. All calculations were performed with the BioShell package.43 Superposition of two BA2930 subunits yields Cα RMSD values of 0.4 Å for both apo- and CoA-bound forms of BA2930. Superposition of each chain of the apo-form on the corresponding chain of the CoA-bound form yields Cα RMSD values ranging from 0.3 to 0.4 Å. These differences can be attributed to mentioned before putative substrate binding lobe. We also observe a similar degree of flexibility in the crystallized mutants (data not shown).