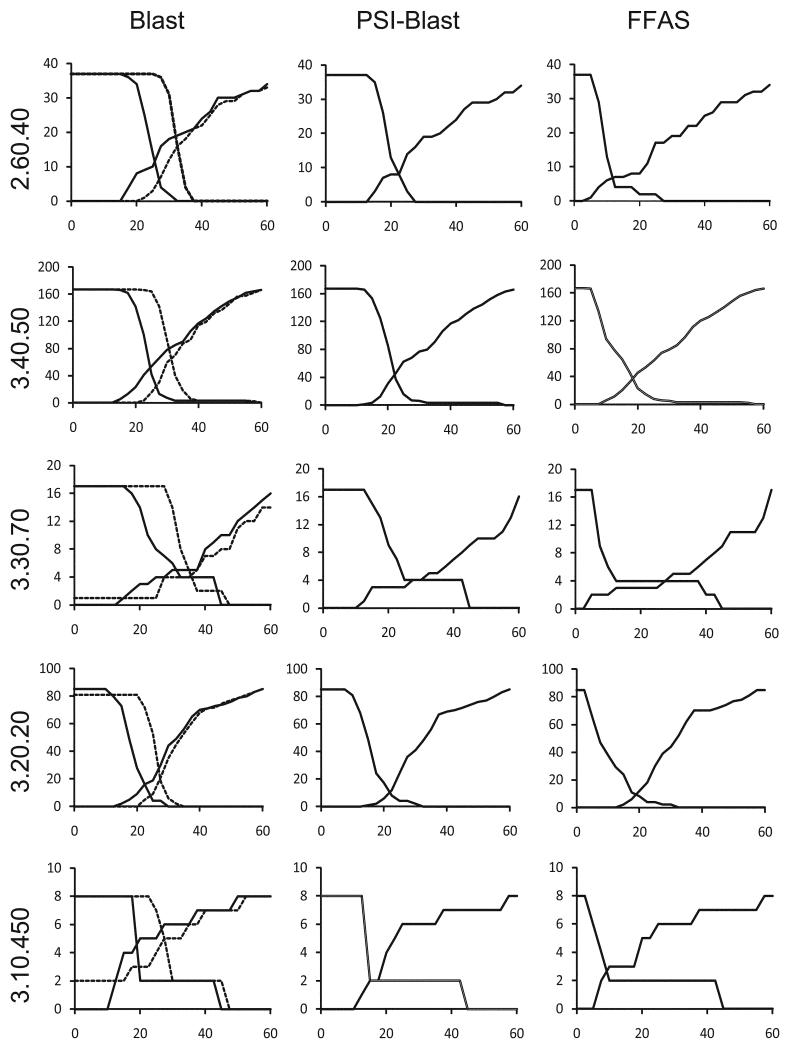
Figure 1.
Predicting structure-based clusters from sequence-based clustering in five large topologies defined in the CATH database. The number of structure-based clusters ‘split’ between different sequence-based clusters and the number of structure-based clusters ‘merged’ into the same sequence-based clusters (Y-axes) are shown as function of sequence identity cutoff used in sequence clustering (X-axes). The number of ‘merged’ structure-based clusters is decreasing with increasing sequence identity cutoff and the number of ‘split’ structure-based clusters is increasing. The intersection of these two curves corresponds to the most accurate prediction of structure-based clusters by sequence-based clusters. Y coordinate of this point provides an assessment of the agreement between sequence-based clusters and structure-based clusters and allows comparison of the accuracy of different sequence clustering algorithms. The corresponding X coordinate gives an optimal sequence identity cutoff for this protein topology (see Methods section for more details). The analysis was performed for Blast, PSI-Blast and FFAS methods. In case of Blast method we tested two ways of normalizing sequence identity. Results obtained with sequence identity normalized by query sequence length (globally normalized sequence identity) and by alignment length (standard sequence identity) are shown as continuous and dashed curves, respectively. The name of CATH topology used in calculations is shown on the left side for each set of graphs.