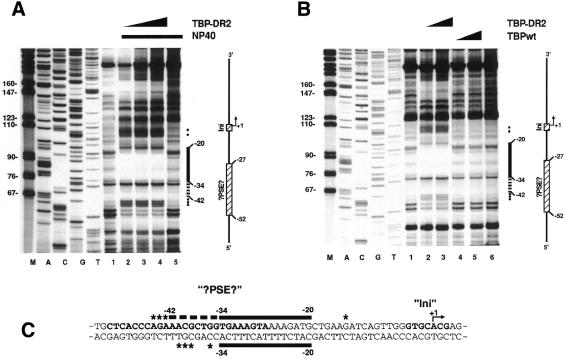
Figure 3.
Footprint of TBP-DR2 and TBPwt on the non-coding strand of the PDR2 promoter. For the investigation of the non-coding strand, a DNA fragment was used, which was amplified by PCR with the M13 sequencing-primer and the 5′-labelled M13 reverse-sequencing-primer using the PDR2-Expression plasmid (Fig. 2B) as template. To be able to identify the region protected by TBP-DR2 a sequencing reaction was conducted in parallel (lanes A, C, G and T). (A) Binding of TBP-DR2 to the PDR2 promoter. Increasing amounts of TBP-DR2 (10, 20 and 40 ng; lanes 2–4) were incubated with the 5′-labelled PDR2 fragment. The control reactions containing no TBP-DR2 (free DNA) are shown in lanes 1 and 5. Lanes 2–5 additionally contained NP-40 at a very low concentration (0.0025%). By this procedure, DNase I protection by TBP-DR2 can be detected with smaller protein quantities and the digestion pattern of DNA is not affected even by higher NP-40 concentrations (data not shown). (B) Comparison of TBPwt and TBP-DR2. Either TBP-DR2 (lane 2, 20 ng; lane 3, 40 ng) or TBPwt (lane 4, 20 ng; lane 5, 40 ng) were incubated as described in (A) but contained no NP-40. Lanes 1 and 6 served as control reactions without protein. (C) Summary of the TBP-DR2 binding to the coding and non-coding strand of the PDR2 promoter sequence. The sequence sections homologous to the hU6 gene (?PSE? and Ini) are written in bold characters. Filled circles and asterisks signify hypersensitive sites in the footprint and schematic representations respectively.