Abstract
Postcopulatory sexual selection is thought to drive the rapid evolution of reproductive tract genes in many animals. Recently, a number of studies have sought to test this hypothesis by examining the effects of mating system variation on the evolutionary rates of reproductive tract genes. Perhaps surprisingly, there is relatively little evidence that reproductive proteins evolve more rapidly in species subject to strong postcopulatory sexual selection. This emerging trend may suggest that other processes, such as host-pathogen interactions, are the main engines of rapid reproductive gene evolution. I suggest that such a conclusion is as yet unwarranted; instead, I propose that more rigorous analytical techniques, as well as multigene and population-based approaches, are required for a full understanding of the consequences of mating system variation for the evolution of reproductive tract genes.
1. Introduction
Over the last two decades, it has become widely appreciated that genes expressed in reproductive tissues, particularly in males, are among the most rapidly evolving genes in the genomes of many organisms. The phenomenon of rapid reproductive tract gene evolution is phylogenetically widespread, having been documented in vertebrates (e.g., [1–4]), invertebrates (e.g., [5, 6]), plants (e.g., [7, 8]), and fungi (e.g., [9]). As such, there is substantial interest in understanding the extent, causes, and consequences of the rapid evolution of reproductive genes.
A number of mechanisms have been proposed to explain the rapid evolution of reproductive tract genes, and the relative contribution of each of these mechanisms likely varies in different taxa. In plants, for example, high within-species diversity of interacting pollen and stigma proteins is vital for the avoidance of self-fertilization [10]. In other species, sexual conflict is thought to play a role in the rapid coevolution of interacting sperm and egg coat proteins, with selection on males favouring sperm that can rapidly penetrate the egg coat and selection on females to avoid polyspermy ([11], but see [12, 13]). Immune processes may also contribute to the rapid evolution of some reproductive proteins: in internally fertilizing animals in particular, copulation is likely to introduce potential pathogens into the female reproductive tract. An arms race between pathogens and hosts could, therefore, underlie selection on female- or male-derived antimicrobial proteins [13–16].
In this perspective, I will focus on the contribution of postcopulatory sexual selection (PCSS) to rapid reproductive protein evolution. Any form of competition amongst ejaculates or biased usage of sperm by females following the act of mating constitutes PCSS, and many such mechanisms have been described (e.g., [17, 18]). A particular form of PCSS, sperm competition, is particularly relevant in the context of the evolution of reproductive tract proteins, and it will be our chief concern in this paper. Sperm competition can occur in internally fertilizing animals if sperm from more than one male are simultaneously present in the reproductive tract of a single female [17, 19, 20]. In such a scenario, competition for fertilization opportunities exerts strong selection on traits that increase a male's paternity at the expense of his opponents. Moreover, females may gain a fitness advantage by biasing their sperm usage towards males of higher quality, more compatible males, males with “sexy” sperm, or males that are more genetically distant (e.g., [21–24]). Such female choice may, in turn, drive sexual conflict, since such female adaptations will decrease the paternity share of some males, resulting in rapid evolution of both male and female traits.
PCSS, therefore, potentially acts on a wide variety of proteins that mediate male-male or male-female postcopulatory interactions. In animals, PCSS may act on sperm and egg surface proteins, as well as seminal fluid proteins and proteins present in the somatic portion of the female reproductive tract. Such proteins have been well studied from both the functional and evolutionary perspectives in a number of model organisms (e.g., [25, 26]). Sperm and egg coat proteins can play important roles in sperm-egg recognition and fusion, and thus may be subject to sexual conflict as described above. Moreover, seminal fluid proteins have been implicated in diverse processes in different animals, including sperm capacitation, sperm storage, and the control of postmating female behaviours, all of which could be subject to PCSS.
PCSS is often cited as a potential, and probable, cause of rapid reproductive protein evolution. Nonetheless, relatively few studies are able to distinguish the action of sexual selection from other processes, such as a host-pathogen arms race, that could also result in the rapid evolution of proteins expressed in reproductive tissues. Recently, a number of researchers have sought to clarify the effects of PCSS by comparing rates of reproductive tract protein evolution in taxa with different mating systems. Here, I critically review the methods used in these efforts, summarize their broad conclusions, and suggest several approaches for making further progress.
2. Rapid Evolution of Reproductive Tract Proteins
Before discussing comparative approaches to reproductive protein evolution, it will be useful to briefly review some of the methods and evidence behind the claim that reproductive tract proteins tend to evolve more rapidly than proteins expressed in other tissues. Most studies have focused on the DNA sequence evolution of protein-coding genes although rapid evolution and/or positive selection has also been documented at the level of protein diversity [27–29] and gene expression [30–32]. Here, I will focus on studies that compare DNA sequences between two or more species, although I will touch on some analyses of within-species sequence variation later on.
Typically, the rate of protein evolution is summarized by the parameter ω, which represents the ratio of dN (the rate of amino-acid changing nucleotide substitutions) to dS (the rate of silent nucleotide substitution). Under the assumption that silent substitutions do not affect fitness, the value of ω reflects the type of selection acting on a gene (or part of a gene). For most genes, average ω across the entire coding region is less than 1, indicating that most amino-acid changes are deleterious and are thus removed by purifying selection. An ω = 1 indicates that amino-acid changes are neutral, while ω > 1 is expected under diversifying (or positive) selection—that is, new amino acids tend to be favoured.
Over the past two decades, many studies have documented evidence for positive selection on selected reproductive tract genes in a variety of species (reviewed in [11, 26, 33–36]). More recently, several papers have shown that large sets of genes expressed in the male reproductive tract have a higher average ω than genes expressed in other tissues. In Drosophila, for example, Haerty and colleagues [5] showed that genes encoding testis- or seminal fluid-specific proteins evolve significantly more rapidly on average than do other classes of gene. Similarly, ω is higher on average for male reproductive tract-specific genes in comparison to other classes of genes in rodents [14, 37] and primates [3]. Interestingly, analogous patterns have generally not been found for genes expressed specifically in the female reproductive tract, with ω for egg, ovary, and/or somatic reproductive tract genes about the same as the genome-wide average in flies and primates [3, 5].
A simple elevation of ω amongst male reproductive tract genes does not necessarily imply the action of diversifying selection; however, this pattern could also be produced by a relaxation of constraint on reproductive tract genes. In order to infer the action of positive selection, it is important to show that ω > 1. Averaged across an entire gene, this criterion is excessively strict, since positive selection likely acts on only a subset of sites. Thus, a number of methods have been developed that allow variation in ω within a gene, and that thereby allow the inference of positive selection (as ω > 1) on a subset of codons. The most popular of these methods is Ziheng Yang's PAML (phylogenetic analysis by maximum likelihood), and a number of other methods build on the models developed by Yang and colleagues [38, 39]. The application of PAML to reproductive tract genes in Drosophila has confirmed that positive selection is at least partly responsible for their elevated ω [5]. In the case of the human/chimpanzee comparison, however, low sequence divergence substantially reduces power to detect positive selection [3], and unavailability of sequence data from multiple species has limited efforts to compare levels of positive selection between reproductive tract genes and nonreproductive tract genes in rodents [14]. More broadly, many studies have documented evidence for positive selection on reproductive tract genes in a wide variety of taxa (see [26, 33] for reviews). However, such studies often select rapidly diverging genes for subsequent PAML analysis (e.g., [37, 40]), and it is, therefore, not possible to contrast the relative impact of positive selection on reproductive-tract and nonreproductive tract genes in these cases.
It is important to note that the methods described in this section assume that rates of evolution are invariant across all lineages considered—that is, there is no variation in ω across the phylogeny of the species studied (Figure 1(a)). While a phylogeny-wide elevation of ω for reproductive tract genes is consistent with the hypothesis that PCSS acts on these genes, these analyses do not exclude other selective mechanisms. Other approaches, such as comparisons of ω between species with different mating systems, will help to clarify the effects of PCSS on rates of molecular evolution.
Figure 1.
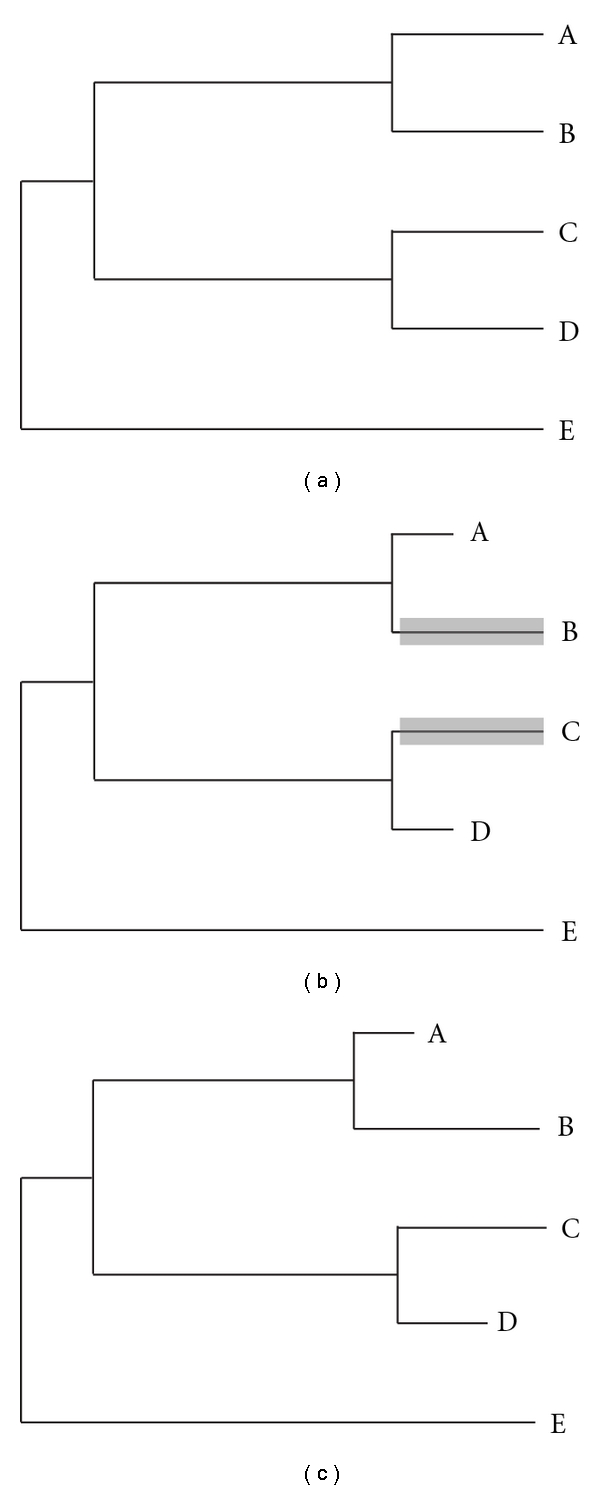
Three types of rate variation. In each panel, branch lengths are proportional to the rate of protein evolution. (a) Amongst the focal species A, B, C, and D, there is no variation in the rate of evolution. (b) Two rate classes. A and D share a rate, and B and C share a faster rate. In the current context, B and C represent polyandrous species, while A and D are monandrous. (c) Species-specific rates. Different rates of evolution are estimated for every lineage.
3. Mating System Variation Explains Morphological Variation in Reproductive Characters
Attempts to associate rates of molecular evolution with variation in the strength of PCSS take their cue from a long history of studies on the relationship between morphological characters and sexual selection. Starting in the late 1970s, a number of studies have examined the relationship between testis size and mating system, primarily in mammals (e.g., [41–49]) but also extending to other animals [50–53]. Many animal taxa show tremendous variation in testis size relative to body size, and it has been hypothesized that large testes are an adaptation to sperm competition. The logic is straightforward: for multiply mating species, such as chimpanzees, it should be in a male's interest to produce many sperm, either to have a numerical advantage in a single bout of sperm competition, or to avoid sperm depletion over the course of multiple matings. By contrast, for species in which females mate with only one male per estrous cycle, for example, gorillas, males need only produce sufficient sperm to ensure fertilization.
Early studies on the relationship between testis size and mating system used a nonphylogenetic approach [46]. Here, log(testes size) is regressed on log(body size), and residuals from this plot are used as a measure of relative testes size, with relatively large testes falling above the regression line and hence having positive residuals, and small testes falling below the regression line with negative residuals (Figure 2(a)). In the primate data presented in Figure 2, for example, it is evident that species in which females mate multiply (circles on Figure 2) have relatively large testes, whereas monandrous species (diamonds on Figure 2) tend to have small testes. However, as Felsenstein [54] pointed out, data from related species are not statistically independent such that regression can yield spurious relationships. Thus, later studies on the relationship between mating system and testis size have used explicitly phylogenetic methods and have generally shown that the association between multiple mating and large testes is, in fact, robust to phylogenetic effects [45, 47, 49, 50]. This relationship can be remarkably strong, with one study in rodents showing that variation in multiple paternity explains between 30% and 50% of variation in testes size [55].
Figure 2.
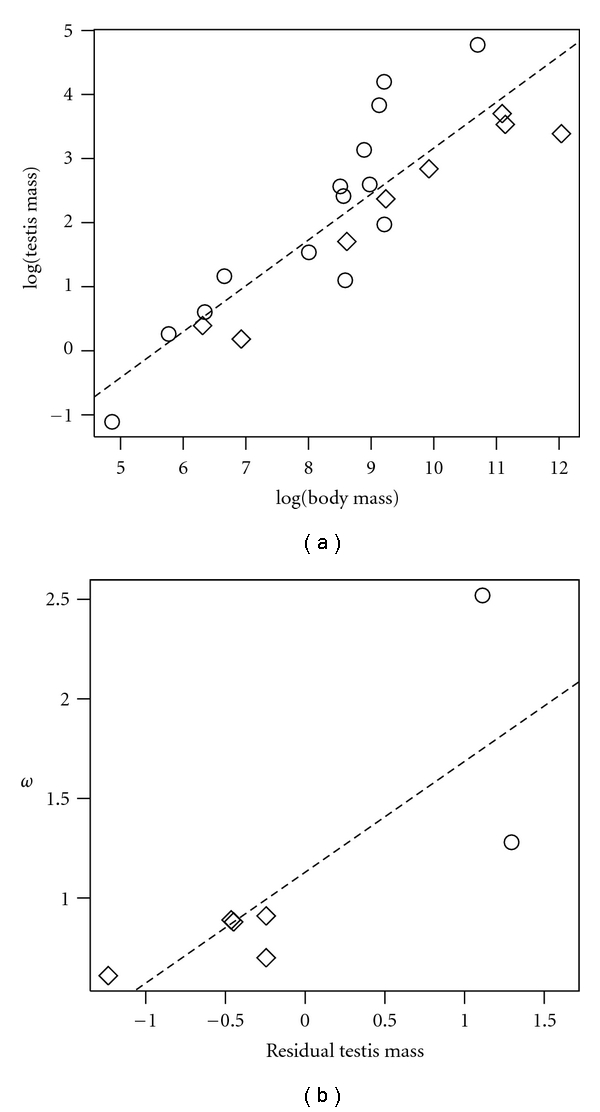
Consequences of sexual selection. (a) Regression of log(testis mass) on log(body mass). Polyandrous species (open circles) tend to have large testes for their body size, and hence fall above the regression line, while monandrous species (diamonds) tend to have relatively small testis. Data are from [47]. (b) Regression of ω on residual testis mass for SEMG2, showing that the rate of protein evolution increases with relative testis size. Estimates of ω are from [56], and testis mass residuals were calculated from panel (a).
The inferred intensity of sperm competition is associated with variation in a myriad of morphological traits beyond testis size. To give just a few examples, divergence in genital morphology is associated with the intensity of sperm competition in many animals [57], and in butterflies female, remating frequency is correlated with both testis size and sperm length [51]. In bats, testis size covaries negatively with brain size [58], probably due to tradeoffs in investment between these two energetically expensive organs. In addition, the presence and/or size of male accessory organs, which produce nonsperm components of the seminal fluid, are associated with sperm competition levels in gobies and in rodents [55, 59]. Similarly, the degree of seminal coagulation correlates with mating system in primates [60]. These results thus suggest that PCSS has evolutionary consequences for a wide range of phenotypic traits.
4. The Comparative Approach in Molecular Evolution
Recently, a number of studies have sought to test the hypothesis that rates of reproductive tract protein evolution, and/or levels of positive selection on these proteins, should increase with the intensity of PCSS or with one of its proxies, such as relative testes mass (see also [61] for an association between mating system and rates of evolution of immunity genes). These attempts represent an important step forward in trying to delineate the causes of the rapid evolution of reproductive tract proteins, rather than merely describe the phenomenon.
Two different methodological approaches have been used to test for a relationship between mating system and rates of molecular evolution. The first method uses categorical descriptions of the mating systems of the species of interest; the specific labels vary somewhat by taxon, but the important distinction is between species with a greater or lesser intensity of sperm competition. Species with low levels of sperm competition include those wherein females mate once per lifetime, species where females mate multiply but where sperm from different males do not overlap in time, and species where females mate with only one male per estrous cycle. Given a distinction between species with high or low sperm competition, the terminal branches of a phylogeny can be labelled by mating system (Figure 1(b)).
The phylogeny, labelled by mating system, can then be used to compare two models of sequence evolution [62]. Under the first model, which serves as the null hypothesis, positive selection (i.e., a subset of codons with ω > 1) is not allowed for any lineage in the phylogeny. Under the second model, the alternative hypothesis, positive selection is allowed on a subset of codons in polyandrous lineages but not in monandrous lineages. Since the null hypothesis is a special case of the alternative, the two models can be compared via a likelihood ratio test. If the data fit the alternative hypothesis better than they do the null hypothesis, then this serves as evidence of an association between mating system and positive selection for the gene under study. This test is implemented in PAML, and corresponds to a comparison between models M2 and M0 described by Zhang and coworkers [62]. It should be noted that this test, as well as the lineage-invariant methods described above, will only detect selection consistently acting on the same codons and is thus likely to miss selection on different codons in different lineages.
This first approach to associating mating system and rates of molecular evolution, which I will refer to as the “discrete” comparative method, has been used by Ramm et al. [1] to study the evolution of seven rodent and two primate reproductive proteins, and by Finn and Civetta [63] to study the evolution of 13 sperm proteins in mammals. Using methods that do not allow variation in ω between lineages, Finn and Civetta found evidence for positive selection on 12 out of 13 genes encoding male expressed ADAM proteins, with positive selection on all 7 sperm surface ADAM proteins. In applying the discrete comparative method, however, only 1 of these 12 genes showed evidence for positive selection specifically along polyandrous lineages, with the 13th sperm-bound ADAM also showing evidence for lineage-specific selection in primates. Thus, the phylogeny-wide signal of positive selection does not appear to be solely, or even chiefly, due to selection in polyandrous species. Similarly, using lineage-invariant methods, Ramm et al. [1] found evidence for positive selection on 5 out of 7 rodent reproductive genes. Using the discrete comparative method, they found evidence for lineage-specific selection on only one of these genes, Svs2, which encodes a copulatory plug protein. Further comparative analyses conducted by Ramm et al. on 2 primate genes will be discussed below.
The second approach for testing for a relationship between mating system and rates of molecular evolution uses continuous measures of the intensity of sperm competition. The most often used metric of PCSS is residual testes mass: given the robustness of the finding that polyandrous species tend to have large testes, residual testes mass should be a good proxy for the intensity of sperm competition. Recently, Herlyn and Zischler [64] and others have proposed that sexual size dimorphism can also be used as a continuous proxy for sperm competition, with less dimorphism indicating stronger sperm competition. The reasoning behind this claim is that in highly dimorphic species, relatively large males will be able to enforce monandry. Finally, estimates of mating frequency—for example, through behavioural observation in primates (e.g., [56]), spermatophore counts in butterflies (e.g., [51]), or genetic analysis of offspring (e.g., [65–67])—can be used as more direct measures of the intensity of sperm competition.
In applying the continuous comparative method (as I will call it), ω is estimated separately for every branch of the phylogenetic tree under consideration (Figure 1(c)). Here, the ω estimate for each branch represents an average across all codons and so is unlikely to significantly exceed 1. Branch-specific estimates of ω are regressed on the continuous measure of sperm competition, and a significant positive relationship (negative in the case of size dimorphism) is interpreted as evidence for an effect of sperm competition on rates of molecular evolution.
Dorus et al. [56] presented the first use of the continuous comparative method on a reproductive protein, in which they studied the molecular evolution of the primate gene SEMG2, which encodes a copulatory plug protein. Lineage-invariant methods find robust evidence for positive selection on this gene, and Dorus et al. found a strong relationship between ω and residual testis size (data reanalyzed in Figure 2(b)), number of mates, and an ordinate rating of semen coagulation.
Several other studies have adopted a similar approach. Herlyn and Zischler [64] found a significantly negative relationship between branch specific ω and sexual size dimorphism in primates for the sperm ligand zonadhesin (ZAN), and Martin-Coello and colleagues [28] obtained a significant association between testis size and promoter divergence for the sperm-specific histone protamine 2. Hurle et al. [68], by contrast, were unable to detect a significant relationship between ω and mating system for any of six reproductive genes in primates (including SEMG2) despite finding evidence for positive selection on 5 of these genes using lineage-invariant methods. The case for an association between mating system and ω for SEMG2 is nonetheless fairly strong: Ramm and colleagues [1] have found evidence for such a relationship using the discrete method for SEMG2 although not for SEMG1 (which encodes a second copulatory plug protein).
Table 1 summarizes the methods and results of studies investigating associations between mating system and rates of molecular evolution, using either the discrete or the continuous comparative methods. Notwithstanding heterogeneity in methods, sample sizes, and taxa, it is striking that very few reproductive genes show evidence for such an association: only 6 genes show evidence for an effect of mating system on rates of protein evolution even though 24 show evidence for positive selection using lineage-invariant methods. This unexpected lack of evidence for an association may indicate that processes other than PCSS underlie the rapid evolution of most reproductive tract proteins, contrary to popular wisdom. However, such a conclusion is likely premature, and I suggest in particular that methods ought to be improved in several ways.
Table 1.
Associations between mating system variation and rates of molecular evolution.
| Study | Taxon | No. genes studied | Method1 | No. genes showing association | No. genes showing positive selection2 |
|---|---|---|---|---|---|
| Dorus et al. 2004 [56] | Primates | 1 | C | 1 | 1 |
| Herlyn and Zischler 2007 [64] | Primates | 1 | C | 1 | 1 |
| Hurle et al. 2007 [68] | Primates | 6 | C | 0 | 5 |
| Ramm et al. 2008 [1] | Rodents | 7 | D | 1 | 5 |
| Primates | 2 | D | 1 | 2 | |
| Martin-Coello et al. 2009 [28] | Rodents | 2 | C | 1 | 1 |
| Finn and Civetta 2010 [63] | Primates | 13 | D | 2 | 12 |
| Total (Unique genes)3 | — | 29 | — | 6 | 24 |
1D = discrete comparative method; C = continuous comparative method.
2Inference of positive selection assuming no variation between lineages.
5. Comparative Methods in Molecular Evolution: Methodological Issues
The statistical methods used to test for associations between mating system and rates of molecular evolution have not been entirely appropriate. The fundamental problem is that estimates of ω on the one hand and morphological or life-history characteristics on the other are different types of data. Phenotypic trait values, measured at the leaves of a phylogenetic tree, are instantaneous measures—that is, they indicate the state of the trait at each leaf now. Estimates of ω, by contrast, are typically an average for the entire branch. Due to this difference in data type, until very recently, there have not been any appropriate methods for detecting covariance between molecular evolutionary rates and phenotypic traits. Felsenstein's method of independent contrasts, for example, is appropriate for instantaneous data, since it explicitly models trait change along the branches of a phylogenetic tree. Attempts to use estimates of ω in an independent contrast framework, thereby conflate an average for an instantaneous measure. Even the discrete comparative method described above runs into problems here, since it assumes that the phenotypic state observed at a leaf has been constant for the entire branch, thus conflating an instantaneous measure for an average. It is not obvious to what extent these methodological concerns will lead to bad inferences, but ideally, we should aim to compare similar types of data in a single statistical framework.
Recently, several model-based methods have been developed for detecting associations between rates of substitution and morphological/life-history variation [69–71]. O'Connor and Mundy [69] and Mayrose and Otto [71] have both developed maximum-likelihood frameworks that simultaneously model molecular evolution and character state evolution. Both formulations use model comparisons to test for a coupling between rates of sequence evolution and character state evolution, in which the character of interest takes on a binary (1/0) value. Under the null model, no such association is present—that is, sequence evolution is independent of the phenotype under consideration. Under the alternative model, rates of molecular evolution vary systematically with the phenotypic character state. A likelihood ratio test can then be used to compare the two models, with a rejection of the null model suggesting an association between phenotypic variation and the rate of molecular evolution. Additional procedures are introduced to handle lineage-specific heterogeneity in rates of sequence evolution that is not associated with the trait of interest (an additional null model in the case of O'Connor and Mundy and parametric bootstrapping in the Mayrose and Otto method).
Currently, both of these methods implement nucleotide substitution models, and hence do not distinguish between synonymous and nonsynonymous substitutions (codon models are used for the latter purpose). As such, associations between molecular evolutionary rates and a phenotypic character are not necessarily specific to changes in protein sequence. Nonetheless, it should be possible to modify either method to use codon models.
Beyond the details of the model implementations, these two methods differ with respect to the formulation of the alternative model. Under the O'Connor and Mundy model, a subset of sites is allowed to evolve in association with the phenotypic trait of interest, with a background substitution rate that is independent of phenotype. By contrast, in the Mayrose and Otto model all sites are assumed to evolve in a phenotype-dependent manner. Thus, the O'Connor and Mundy approach may be more suitable when selection drives the evolution of some, but not all, sites in a lineage specific manner, whereas the Mayrose and Otto method is best suited for detecting lineage-specific mutational effects that influence all sites.
Lartillot and Poujol [70] have developed a Bayesian method, CoEvol, for detecting associations between rates of molecular evolution and rates of change in phenotypic/life history traits. Unlike the O'Connor and Mundy [69] and Mayrose and Otto [71] methods, CoEvol considers continuous phenotypic characters, and implements a codon model of sequence evolution rather than a nucleotide model. CoEvol estimates a covariance matrix for the rates of change of dS, ω, and one or more morphological/life history characters. A high posterior probability associated with covariance between the rate of change in dS or ω on the one hand, and the rate of change of a phenotypic character on the other, serves as evidence for coupling between the molecular and phenotypic processes. Importantly, separate estimates of covariance for dS and ω should allow one to distinguish mutational effects (via covariance with dS) from selective effects (via covariance with ω).
The phylogenetic methods just described have each been applied to different datasets: Mayrose and Otto [71], for example, detected an effect of habitat salinity on the rate of molecular evolution in daphniids, and Lartillot and Poujol [70] detected negative associations between dS and mass and longevity in mammals. Only O'Connor and Mundy [69] have applied their method to reproductive proteins, specifically the primate seminal proteins SEMG1, SEMG2, and Zonadhesin (ZAN) as well as two genes not expected to be subject to sexual selection, PI3 and CYTB. Using a binary mating system classification (multiply mating versus single mating), they found a significant association between substitution rate and phenotypic state for SEMG2, but not for the other four loci. Thus, their analyses are consistent with previous studies for SEMG2, but not for ZAN (recall that Herlyn and Zischler [64] reported a negative association between sexual size dimorphism and ω for the ZAN locus). The discrepancy in the case of ZAN could reflect differences in the way PCSS is measured (binary classification versus sexual size dimorphism), lack of power in the the O'Connor and Mundy method, or may suggest that the previously reported correlation between ω and dimorphism is a methodological artifact. Further studies will be required to distinguish these possibilities.
These new statistical approaches should prove to be powerful tools for investigating the consequences of mating system variation for substitution rates, since they will lend more confidence to inferences concerning associations (or lack thereof) between mating system and rates of molecular evolution. It will be particularly important to investigate the statistical properties of these methods (power, false positive rates under different conditions, etc.) in order to better understand when and how each should be used.
6. Inconsistency in the Targets of Selection
To date, most studies on the relationship between mating system and the molecular evolution of reproductive proteins have focused on one or a few genes. Thus, in order to detect an association, PCSS must act on the same gene in most lineages. While this may be true in some cases (e.g., SEMG2), it is not a foregone conclusion that the same genes will be subject to PCSS in different species. Indeed, the rapid turnover of reproductive tract genes in some species (e.g., Drosophila—[5, 72]), as well as observations of lineage specific positive selection on reproductive proteins [37, 63], suggests that there may be variation in the targets of selection between lineages.
If PCSS operates on different loci in different species, then it may be fruitful to compare average rates of evolution across many reproductive genes between taxa with different mating systems. Indeed, a handful of studies have done this and, in general, confirm the prediction that average rates of evolution are higher in taxa with higher levels of PCSS. For example, remating rates in the repleta group of the fruitfly genus Drosophila are substantially higher than remating rates in the melanogaster group (e.g., [73, 74]) such that the intensity of PCSS is presumably higher for repleta group species. A number of recent studies have reported that average ω and rates of gene duplication are higher for both male and female reproductive tract genes in the repleta group than in the melanogaster group [75–78], as predicted if PCSS drives reproductive tract gene evolution. Similarly, I recently reported that rates of evolution are higher for testis-specific proteins in highly polyandrous chimpanzees than in humans, where historical levels of polyandry are thought to be more modest [3]. Work in additional taxa will be required to confirm these patterns (e.g., [79]), but these first studies suggest that we should not necessarily expect a strong signal of PCSS when looking at single genes alone.
In addition to this “many-gene” approach, molecular population genetic studies of candidate genes, rather than between-species comparisons, should provide complementary data on the effects of mating system on the evolution of genes encoding reproductive proteins. Here, candidate genes would be sequenced in multiple individuals from one or more species with strong PCSS, as well as in multiple individuals from one or more species with no or weak PCSS. A wide variety of statistical tests are available for inferring recent selection from such data [80, 81], and application of these tests would allow gene-by-gene tests of the prediction that positive selection should act in multiply mating species but not in monandrous species. The advantage of this approach is that selection need only have acted in a single species to detect a signal, whereas selection must be fairly consistent across a phylogeny to detect positive selection in between-species comparisons. Population genetic surveys in primates have proved informative with respect to semenogelin (SEMG1 and SEMG2) evolution, for example. In chimpanzees, which are highly polyandrous, SEMG1 shows strong evidence for recent positive selection [82] and an increase in protein length owing to a repeat expansion [83]. In gorillas, by contrast, both SEMG1 and SEMG2 carry multiple loss-of-function mutations, consistent with a loss of constraint due to the highly monandrous mating-system of this species [82, 83].
7. Summary and Conclusions
Despite widespread evidence for the rapid evolution of, and positive selection on, genes encoding reproductive tract proteins, comparative studies have had relatively little success in associating rates of protein evolution with the strength of PCSS. Only a handful of individual genes show robust evidence for accelerated evolution in polyandrous lineages in comparison to monandrous ones. This lack of evidence for a relationship between mating system and rates of molecular evolution may indicate that processes other than PCSS, such as immune interactions, drive the evolution of many reproductive tract proteins. I have suggested, however, that new analytical methods will add rigour to attempts to delineate the causes of rapid reproductive tract protein evolution. Moreover, if selection acts on different genes in different lineages, then a combination of multigene comparative studies, as well as population genetic studies, should prove useful.
Acknowledgments
The author thanks three anonymous reviewers, and the editors of this special issue for helpful comments on this paper. The author acknowledges funding support from NSERC, CIHR, and the Banting Fellowship program.
References
- 1.Ramm SA, Oliver PL, Ponting CP, Stockley P, Emes RD. Sexual selection and the adaptive evolution of mammalian ejaculate proteins. Molecular Biology and Evolution. 2008;25(1):207–219. doi: 10.1093/molbev/msm242. [DOI] [PubMed] [Google Scholar]
- 2.Turner LM, Hoekstra HE. Adaptive evolution of fertilization proteins within a genus: variation in ZP2 and ZP3 in deer mice (Peromyscus) Molecular Biology and Evolution. 2006;23(9):1656–1669. doi: 10.1093/molbev/msl035. [DOI] [PubMed] [Google Scholar]
- 3.Wong A. Testing the effects of mating system variation on rates of molecular evolution in primates. Evolution. 2010;64(9):2779–2785. doi: 10.1111/j.1558-5646.2010.01038.x. [DOI] [PubMed] [Google Scholar]
- 4.Clark NL, Swanson WJ. Pervasive adaptive evolution in primate seminal proteins. PLoS Genetics. 2005;1(3, article e35) doi: 10.1371/journal.pgen.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haerty W, Jagadeeshan S, Kulathinal RJ, et al. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177(3):1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark NL, Gasper J, Sekino M, Springer SA, Aquadro CF, Swanson WJ. Coevolution of interacting fertilization proteins. PLoS Genetics. 2009;5(7) doi: 10.1371/journal.pgen.1000570. Article ID e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takebayashi N, Brewer PB, Newbigin ED, Uyenoyama MK. Patterns of variation within self-incompatibility loci. Molecular Biology and Evolution. 2003;20(11):1778–1794. doi: 10.1093/molbev/msg209. [DOI] [PubMed] [Google Scholar]
- 8.Sato K, Nishio T, Kimura R, et al. Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics. 2002;162(2):931–940. doi: 10.1093/genetics/162.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wik L, Karlsson M, Johannesson H. The evolutionary trajectory of the mating-type (mat) genes in Neurospora relates to reproductive behavior of taxa. BMC Evolutionary Biology. 2008;8(1):p. 109. doi: 10.1186/1471-2148-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takayama S, Isogai A. Self-incompatibility in plants. Annual Review of Plant Biology. 2005;56:467–489. doi: 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- 11.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nature Reviews Genetics. 2002;3(2):137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 12.Calkins JD, El-Hinn D, Swanson WJ. Adaptive evolution in an avian reproductive protein: ZP3. Journal of Molecular Evolution. 2007;65(5):555–563. doi: 10.1007/s00239-007-9034-8. [DOI] [PubMed] [Google Scholar]
- 13.Berlin S, Qu L, Ellegren H. Adaptive evolution of gamete-recognition proteins in birds. Journal of Molecular Evolution. 2008;67(5):488–496. doi: 10.1007/s00239-008-9165-6. [DOI] [PubMed] [Google Scholar]
- 14.Dean MD, Clark NL, Findlay GD, et al. Proteomics and comparative genomic investigations reveal heterogeneity in evolutionary rate of male reproductive proteins in mice (Mus domesticus) Molecular Biology and Evolution. 2009;26(8):1733–1743. doi: 10.1093/molbev/msp094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Good JM, Nachman MW. Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Molecular Biology and Evolution. 2005;22(4):1044–1052. doi: 10.1093/molbev/msi087. [DOI] [PubMed] [Google Scholar]
- 16.Lawniczak MKN, Barnes AI, Linklater JR, Boone JM, Wigby S, Chapman T. Mating and immunity in invertebrates. Trends in Ecology and Evolution. 2007;22(1):48–55. doi: 10.1016/j.tree.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Parker GA. Sperm competition and its evolutionary consequences in the insects. Biological Reviews. 1970;45(4):525–567. [Google Scholar]
- 18.Eberhard WG. Female Control: Sexual Selection by Cryptic Female Choice. Princeton, NJ, USA: Princeton University Press; 1996. [Google Scholar]
- 19.Parker GA. Sexual conflict over mating and fertilization: an overview. Philosophical Transactions of the Royal Society of London: Series B. 2006;361(1466):235–259. doi: 10.1098/rstb.2005.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birkhead TR. How stupid not to have thought of that: post-copulatory sexual selection. Journal of Zoology. 2010;281(2):78–93. [Google Scholar]
- 21.Keller L, Reeve HK. Why do females mate with multiple males—the sexually selected sperm hypothesis. Advances in the Study of Behavior. 1995;24:291–315. [Google Scholar]
- 22.Kokko H, Jennions MD, Brooks R. Unifying and testing models of sexual selection. Annual Review of Ecology, Evolution, and Systematics. 2006;37(1):43–66. [Google Scholar]
- 23.Andersson M, Simmons LW. Sexual selection and mate choice. Trends in Ecology and Evolution. 2006;21(6):296–302. doi: 10.1016/j.tree.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 24.Simmons LW. The evolution of polyandry: sperm competition, sperm selection, and offspring viability. Annual Review of Ecology, Evolution, and Systematics. 2005;36(1):125–146. [Google Scholar]
- 25.Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF. Insect seminal fluid proteins: identification and function. Annual Review of Entomology. 2011;56:21–40. doi: 10.1146/annurev-ento-120709-144823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panhuis TM, Clark NL, W. J. S. Rapid evolution of reproductive proteins in abalone and Drosophila. Philosophical Transactions of the Royal Society of London: Series B. 2006;361(1466):261–268. doi: 10.1098/rstb.2005.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Civetta A, Singh RS. High divergence of reproductive tract proteins and their association with postzygotic reproductive isolation in Drosophila melanogaster and Drosophila virilis species. Journal of Molecular Evolution. 1995;41(6):1085–1095. doi: 10.1007/BF00173190. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Coello J, Dopazo H, Arbiza L, Ausió J, Roldan ERS, Gomendio M. Sexual selection drives weak positive selection in protamine genes and high promoter divergence, enhancing sperm competitiveness. Proceedings of the Royal Society B. 2009;276(1666):2427–2436. doi: 10.1098/rspb.2009.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramm SA, McDonald L, Hurst JL, Beynon RJ, Stockley P. Comparative proteomics reveals evidence for evolutionary diversification of rodent seminal fluid and its functional significance in sperm competition. Molecular Biology and Evolution. 2009;26(1):189–198. doi: 10.1093/molbev/msn237. [DOI] [PubMed] [Google Scholar]
- 30.Voolstra C, Tautz D, Farbrother P, Eichinger L, Harr B. Contrasting evolution of expression differences in the testis between species and subspecies of the house mouse. Genome Research. 2007;17(1):42–49. doi: 10.1101/gr.5683806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Molecular Biology and Evolution. 2004;21(7):1308–1317. doi: 10.1093/molbev/msh128. [DOI] [PubMed] [Google Scholar]
- 32.Meiklejohn CD, Parsch J, Ranz JM, Hartl DL. Rapid evolution of male-biased gene expression in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(17):9894–9899. doi: 10.1073/pnas.1630690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131(1):11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- 34.Chapman T. The soup in my fly: evolution, form and function of seminal fluid proteins. PLoS Biology. 2008;6(7, article e179) doi: 10.1371/journal.pbio.0060179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Civetta A. Shall we dance or shall we fight? using DNA sequence data to untangle controversies surrounding sexual selection. Genome. 2003;46(6):925–929. doi: 10.1139/g03-109. [DOI] [PubMed] [Google Scholar]
- 36.Turner LM, Hoekstra HE. Causes and consequences of the evolution of reproductive proteins. International Journal of Developmental Biology. 2008;52(5-6):769–780. doi: 10.1387/ijdb.082577lt. [DOI] [PubMed] [Google Scholar]
- 37.Turner LM, Chuong EB, Hoekstra HE. Comparative analysis of testis protein evolution in rodents. Genetics. 2008;179(4):2075–2089. doi: 10.1534/genetics.107.085902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Molecular Biology and Evolution. 1994;11(5):725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, Nielsen R, Goldman N, Pedersen AMK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155(1):431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swanson WJ, Wong A, Wolfner MF, Aquadro CF. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics. 2004;168(3):1457–1465. doi: 10.1534/genetics.104.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Short RV. Sexual selection and its component parts, somatic and genital selection, as illustrated by man and the great apes. Advances in the Study of Behavior. 1979;9:131–158. [Google Scholar]
- 42.Rose RW, Nevison CM, Dixson AF. Testes weight, body weight and mating systems in marsupials and monotremes. Journal of Zoology. 1997;243(3):523–531. [Google Scholar]
- 43.Møller AP. Ejaculate quality, testes size and sperm competition in primates. Journal of Human Evolution. 1988;17(5):479–488. [Google Scholar]
- 44.Kenagy G, Trombulak S. Size and function of mammalian testes in relation to body size. Journal of Mammalogy. 1986 ;67(1):1–22. [Google Scholar]
- 45.Hosken DJ. Testes mass in megachiropteran bats varies in accordance with sperm competition theory. Behavioral Ecology and Sociobiology. 1998;44(3):169–177. [Google Scholar]
- 46.Harcourt AH, Harvey PH, Larson SG, Short RV. Testis weight, body weight and breeding system in primates. Nature. 1981;293(5827):55–57. doi: 10.1038/293055a0. [DOI] [PubMed] [Google Scholar]
- 47.Harcourt AH, Purvis A, Liles L. Sperm competition: mating system, not breeding season, affects testes size of primates. Functional Ecology. 1995;9(3):468–476. [Google Scholar]
- 48.Arnqvist G. Comparative evidence for the evolution of genitalia by sexual selection. Nature. 1998;393(6687):784–786. [Google Scholar]
- 49.Iossa G, Soulsbury CD, Baker PJ, Harris S. Sperm competition and the evolution of testes size in terrestrial mammalian carnivores. Functional Ecology. 2008;22(4):655–662. [Google Scholar]
- 50.Stockley P, Gage MJG, Parker GA, Møller AP. Sperm competition in fishes: the evolution of testis size and ejaculate characteristics. The American Naturalist. 1997;149(5):933–954. doi: 10.1086/286031. [DOI] [PubMed] [Google Scholar]
- 51.Gage MJG. Associations between body size, mating pattern, testis size and sperm lengths across butterflies. Proceedings of the Royal Society B. 1994;258(1353):247–254. [Google Scholar]
- 52.Emerson SB. Testis size variation in frogs: testing the alternatives. Behavioral Ecology and Sociobiology. 1997;41(4):227–235. [Google Scholar]
- 53.Calhim S, Birkhead TR. Testes size in birds: quality versus quantity—assumptions, errors, and estimates. Behavioral Ecology. 2007;18(1):271–275. [Google Scholar]
- 54.Felsenstein J. Phylogenies and the comparative method. The American Naturalist. 1985;125(1):1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- 55.Ramm SA, Parker GA, Stockley P. Sperm competition and the evolution of male reproductive anatomy in rodents. Proceedings of the Royal Society B. 2005;272(1566):949–955. doi: 10.1098/rspb.2004.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT. Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nature Genetics. 2004;36(12):1326–1329. doi: 10.1038/ng1471. [DOI] [PubMed] [Google Scholar]
- 57.Hosken DJ, Stockley P. Sexual selection and genital evolution. Trends in Ecology and Evolution. 2004;19(2):87–93. doi: 10.1016/j.tree.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 58.Pitnick S, Jones KE, Wilkinson GS. Mating system and brain size in bats. Proceedings of the Royal Society B. 2006;273(1587):719–724. doi: 10.1098/rspb.2005.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mazzoldi C, Petersen CW, Rasotto MB. The influence of mating system on seminal vesicle variability among gobies (Teleostei, Gobiidae) Journal of Zoological Systematics and Evolutionary Research. 2005;43(4):307–314. [Google Scholar]
- 60.Dixson AF, Anderson MJ. Sexual selection, seminal coagulation and copulatory plug formation in primates. Folia Primatologica. 2002;73(2–3):63–69. doi: 10.1159/000064784. [DOI] [PubMed] [Google Scholar]
- 61.Wlasiuk G, Nachman MW. Promiscuity and the rate of molecular evolution at primate immunity genes. Evolution. 2010;64(8):2204–2220. doi: 10.1111/j.1558-5646.2010.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Molecular Biology and Evolution. 2005;22(12):2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 63.Finn S, Civetta A. Sexual selection and the molecular evolution of ADAM proteins. Journal of Molecular Evolution. 2010;71(3):231–240. doi: 10.1007/s00239-010-9382-7. [DOI] [PubMed] [Google Scholar]
- 64.Herlyn H, Zischler H. Sequence evolution of the sperm ligand zonadhesin correlates negatively with body weight dimorphism in primates. Evolution. 2007;61(2):289–298. doi: 10.1111/j.1558-5646.2007.00035.x. [DOI] [PubMed] [Google Scholar]
- 65.Bretman A, Tregenza T. Measuring polyandry in wild populations: a case study using promiscuous crickets. Molecular Ecology. 2005;14(7):2169–2179. doi: 10.1111/j.1365-294X.2005.02556.x. [DOI] [PubMed] [Google Scholar]
- 66.Dean MD, Ardlie KG, Nachman MW. The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus) Molecular Ecology. 2006;15(13):4141–4151. doi: 10.1111/j.1365-294X.2006.03068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Griffith SC, Owens IPF, Thuman KA. Extra pair paternity in birds: a review of interspecific variation and adaptive function. Molecular Ecology. 2002;11(11):2195–2212. doi: 10.1046/j.1365-294x.2002.01613.x. [DOI] [PubMed] [Google Scholar]
- 68.Hurle B, Swanson W, Green ED. Comparative sequence analyses reveal rapid and divergent evolutionary changes of the WFDC locus in the primate lineage. Genome Research. 2007;17(3):276–286. doi: 10.1101/gr.6004607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Connor TD, Mundy NI. Genotype-phenotype associations: substitution models to detect evolutionary associations between phenotypic variables and genotypic evolutionary rate. Bioinformatics. 2009;25(12):i94–i100. doi: 10.1093/bioinformatics/btp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lartillot N, Poujol R. A phylogenetic model for investigating correlated evolution of substitution rates and continuous phenotypic characters. Molecular Biology and Evolution. 2010;28(1):729–744. doi: 10.1093/molbev/msq244. [DOI] [PubMed] [Google Scholar]
- 71.Mayrose I, Otto SP. A likelihood method for detecting trait-dependent shifts in the rate of molecular evolution. Molecular Biology and Evolution. 2010;28(1):759–770. doi: 10.1093/molbev/msq263. [DOI] [PubMed] [Google Scholar]
- 72.Holloway AK, Begun DJ. Molecular evolution and population genetics of duplicated accessory gland protein genes in Drosophila. Molecular Biology and Evolution. 2004;21(9):1625–1628. doi: 10.1093/molbev/msh195. [DOI] [PubMed] [Google Scholar]
- 73.Markow T. Evolution of Drosophila mating systems. Evolutionary Biology. 1996;29:73–106. [Google Scholar]
- 74.Markow TA, O’Grady PM. Evolutionary genetics of reproductive behavior in Drosophila: connecting the dots. Annual Review of Genetics. 2005;39:263–291. doi: 10.1146/annurev.genet.39.073003.112454. [DOI] [PubMed] [Google Scholar]
- 75.Kelleher ES, Swanson WJ, Markow TA. Gene duplication and adaptive evolution of digestive proteases in Drosophila arizonae female reproductive tracts. PLoS Genetics. 2007;3(8, article e148) doi: 10.1371/journal.pgen.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Almeida FC, Desalle R. Orthology, function and evolution of accessory gland proteins in the Drosophila repleta group. Genetics. 2009;181(1):235–245. doi: 10.1534/genetics.108.096263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wagstaff BJ, Begun DJ. Adaptive evolution of recently duplicated accessory gland protein genes in desert Drosophila. Genetics. 2007;177(2):1023–1030. doi: 10.1534/genetics.107.077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wagstaff BJ, Begun DJ. Molecular population genetics of accessory gland protein genes and testis-expressed genes in Drosophila mojavensis and D. arizonae. Genetics. 2005;171(3):1083–1101. doi: 10.1534/genetics.105.043372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walters JR, Harrison RG. Combined EST and proteomic analysis identifies rapidly evolving seminal fluid proteins in Heliconius butterflies. Molecular Biology and Evolution. 2010;27(9):2000–2013. doi: 10.1093/molbev/msq092. [DOI] [PubMed] [Google Scholar]
- 80.Nielsen R. Molecular signatures of natural selection. Annual Review of Genetics. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 81.Jensen JD, Wong A, Aquadro CF. Approaches for identifying targets of positive selection. Trends in Genetics. 2007;23(11):568–577. doi: 10.1016/j.tig.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 82.Kingan SB, Tatar M, Rand DM. Reduced polymorphism in the chimpanzee semen coagulating protein, semenogelin I. Journal of Molecular Evolution. 2003;57(2):159–169. doi: 10.1007/s00239-002-2463-0. [DOI] [PubMed] [Google Scholar]
- 83.Jensen-Seaman MI, Li WH. Evolution of the hominoid semenogelin genes, the major proteins of ejaculated semen. Journal of Molecular Evolution. 2003;57(3):261–270. doi: 10.1007/s00239-003-2474-x. [DOI] [PubMed] [Google Scholar]


