Table 1. Model parameter values.
| description | parameter | value |
| cell-cell adhesion energy density |
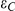
|
600 µN/m |
| cell-substrate adhesion energy/area |
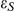
|
200 µN/m |
| Poisson ratio |

|
⅓ |
| Young modulus |
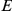
|
1 kPa |
| bulk compression modulus |
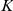
|
1 kPa |
| cell-cell friction constant |
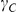
|
2.8 109 Ns/m3 |
| cell-substrate friction constant |
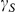
|
2.8 109 Ns/m3 |
| cell radius friction constant |
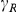
|
173 109 Ns/m3 |
| cell viscous friction constant |

|
0.04 Ns/m |
| number of podia-related update rate |
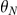
|
1440 1/d |
| independent podium update rate |
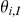
|
[2…0] 1/d |
| number of podia offset value |
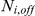
|
[3.5…2.5] |
| podium angle scaling constant |
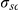
|
3.0 rad |
| protrusion force |
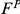
|
2.5 nN |
| podium-substrate friction constant |
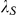
|
0.07 Ns/m |
| podium spring constant |
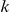
|
0.1 nN/µm |
| initial radius of cell body |
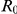
|
4.75 µm |
| target volume increment per time |
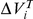
|
[1200…0] µm3/d |
| average number of volume increments per time |
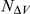
|
20 1/d |
| distance for nearest neighbor classification |
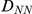
|
5 µm |
| number of nearest neighbors – proliferation |
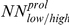
|
[4…5] |
| number of nearest neighbors – podia |
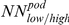
|
[1…3] |
Numbers given in square brackets refer to values at low and high cell density, respectively, i.e. value at [low density … high density]. 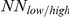 define the low and high cell density regimes, respectively (see subsection G). For parameter values related to the spherical cell bodies refer to our previous publications [34], [49], [50]. The number of podia-related update rate corresponds to the podia generation time of about one minute as measured for eukaryotic cells [53]. The maximum protrusion force of podia ranges between 0.5 and 10
define the low and high cell density regimes, respectively (see subsection G). For parameter values related to the spherical cell bodies refer to our previous publications [34], [49], [50]. The number of podia-related update rate corresponds to the podia generation time of about one minute as measured for eukaryotic cells [53]. The maximum protrusion force of podia ranges between 0.5 and 10  depending on cell type [94], [95].
depending on cell type [94], [95].
