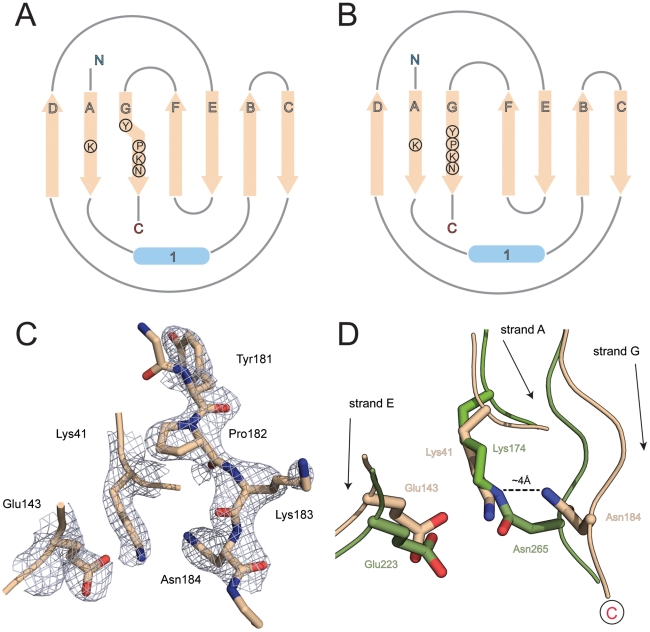
Figure 2. β-sheet topology and isopeptide bond formation.
A) Topological representation of domain D1 of RrgB. The final β-strand G carries the YPKN pilin motif residues, which include the lysine used for ligation to the next subunit. In RrgB D1, strand G diverges from the N-terminal strand A at the proline residue of this motif, transferring its hydrogen bonding to strand F. B) In a conventional CnaB domain, which comprises a 3-stranded β-sheet D-A-G and a 4-stranded sheet F-E-B-C and strand G is hydrogen bonded to strand A along its whole length, allowing an isopeptide bond to be formed between a lysine in strand A and an asparagine in strand G. C) Electron density for the YPKN motif (residues 181–184) in β-strand G of domain D1 together with that for the other two residues that could potentially participate in internal isopeptide bond formation, Lys41 and Glu143. The electron density is from a 2Fo – Fc map contoured at 0.38 eÅ−3 (1.5σ). D) Superposition of strands A and E of RrgB D1 (wheat) on to the equivalent strands of domain D2 of BcpA from B. cereus (green) showing that an isopeptide bond such as that present in BcpA between Lys174 and Asn265 cannot occur in RrgB D1 because the divergence of strand G in RrgB D1 pulls Asn184∼4 Å further away from the lysine (Lys41) in strand A.