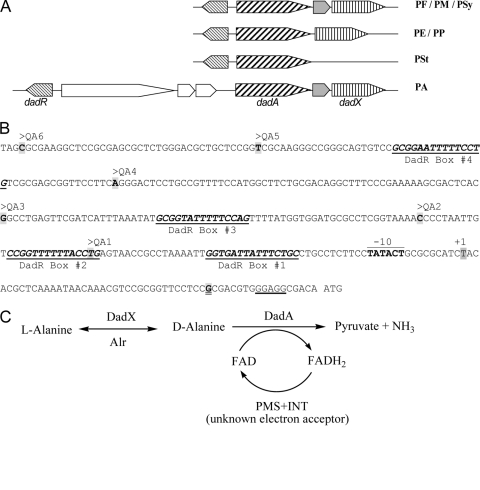
Fig. 1.
(A) Gene organization of the dadRAX locus in Pseudomonas. Orthologues of dadRAX are shown in the same shade. PA, P. aeruginosa; PE, P. entomophila; PF, P. fluorescens; PM, P. mendocina; PP, P. putida; PSt, P. stutzeri; PSy, P. syringae. (B) The nucleotide sequence of the dadA regulatory region in P. aeruginosa PAO1. The −10 element and the transcriptional initiation site (+1) of the dadA promoter as described by Boulette et al. (1) are labeled, and the ribosome binding site in front of the ATG initiation codon of dadA is underlined. The 5′ ends of probes QA1 to -6 are labeled accordingly, the common 3′ end of these probes is highlighted and double underlined, and four putative DadR binding sites are numbered and underlined. (C) Proposed functions of the d-amino acid dehydrogenase DadA and the amino acid racemases DadX and Alr in alanine catabolism. PMS and INT are artificial electron acceptors in the assays for DadA as described in Materials and Methods.