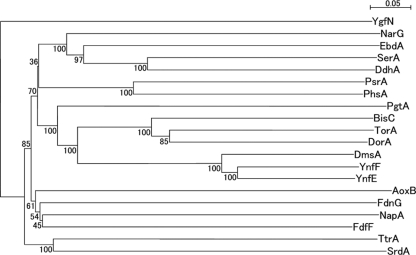
Fig. 5.
Neighbor-joining phylogenetic tree of representative catalytic subunits, including SrdA. Multiple-alignment and phylogenetic analyses were performed using the ClustalW program. The tree was generated using NJplot. Five percent substitution of the sequence is indicated by a bar. Bootstrap values were calculated from 100 replicates. All protein sequences were obtained from the Swiss-Prot database: YgfN, putative hypoxanthine oxidase in Escherichia coli K-12; FdnG, formate dehydrogenase in E. coli K-12; AoxB, arsenate oxidase in Alcaligenes faecalis; NapA, periplasmic nitrate reductase in E. coli K-12; FdhF, formate dehydrogenase in E. coli K-12; PsrA, polysulfide reductase in Wolinella succinogenes; PhsA, thiosulfate reductase in Salmonella enterica serovar Typhimurium; NarG, nitrate reductase in E. coli K-12; EbdA, ethylbenzene dehydrogenase in Azoarcus sp. strain EB1; SerA, selenate reductase in Thauera selenatis; DdhA, dimethylsulfide dehydrogenase in Rhodovulum sulfidophilum; PgtL, pyrogallol hydroxytransferase in Pelobacter acidigallici; BisC, biotin sulfoxide reductase in E. coli K-12; TorA, trimethylamine-N-oxide reductase in E. coli K-12; DorA, DMSO/trimethylamine N-oxide reductase in Rhodobacter capsulatus; DmsA, DMSO reductase in E. coli K-12; YnfF, probable DMSO reductase in E. coli K-12; YnfE, putative DMSO reductase in E. coli K-12; TtrA, tetrathionate reductase in S. enterica serovar Typhimurium; SrdA, selenate reductase in Bacillus selenatarsenatis SF-1.