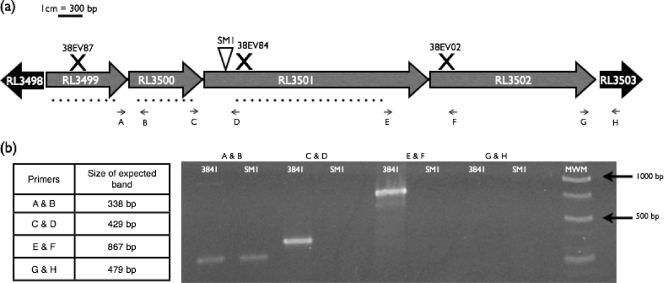
Fig. 1.
(a) Schematic of the RL3499-RL3502 operon region. Gene annotations were obtained from the Rhizobase database (Kazusa DNA Research Institute; http://bacteria.kazusa.or.jp/rhizobase/). The triangle indicates the transposon insertion site, X's indicate the locations of disruption of insertional mutants, and dotted lines indicate regions deleted in the nonpolar mutants. Arrows indicate primer binding sites. (b) RT-PCR of the wild-type strain and the SM1 mutant to determine operon structure. MWM, molecular weight marker (GeneRuler 1-kb DNA ladder; Fermentas). Controls with no reverse transcriptase did not yield amplification products, indicating that no contaminating DNA was present in the RNA samples, and all sets of primers were confirmed to amplify the amplicon of the appropriate size by using standard PCR and genomic DNA from wild-type strain 3841 as the template (data not shown).