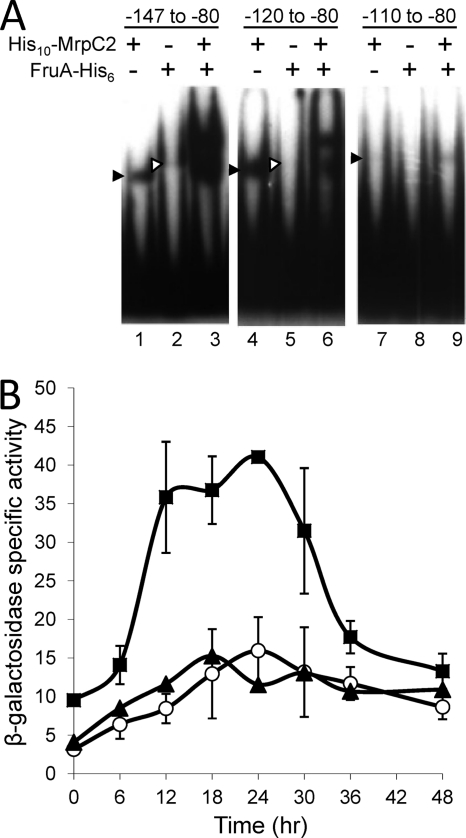
Fig. 4.
MrpC2 and FruA bind to an upstream negative regulatory element. (A) Binding of MrpC2 and FruA to site 1. EMSAs with 32P-labeled fmgE DNA (2 nM) spanning from the indicated 5′ ends to position −80 and His10-MrpC2 (1 μM), FruA-His6 (3 μM), or both His10-MrpC2 (1 μM) and FruA-His6 (3 μM), as indicated. The filled arrowhead points to the shifted complex produced by His10-MrpC2 alone, and the open arrowhead points to the complex produced by FruA-His6 alone. (B) Effects of 5′-end deletions on developmental fmgE-lacZ expression. β-Galactosidase specific activity during development was measured for lacZ fused to fmgE spanning from positions −150 to +50 (▴), −120 to +50 (○), or −110 to +50 (■). The units of activity are nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. Points show the average values of two or three transformants, and each error bars depicts 1 standard deviation of the data.