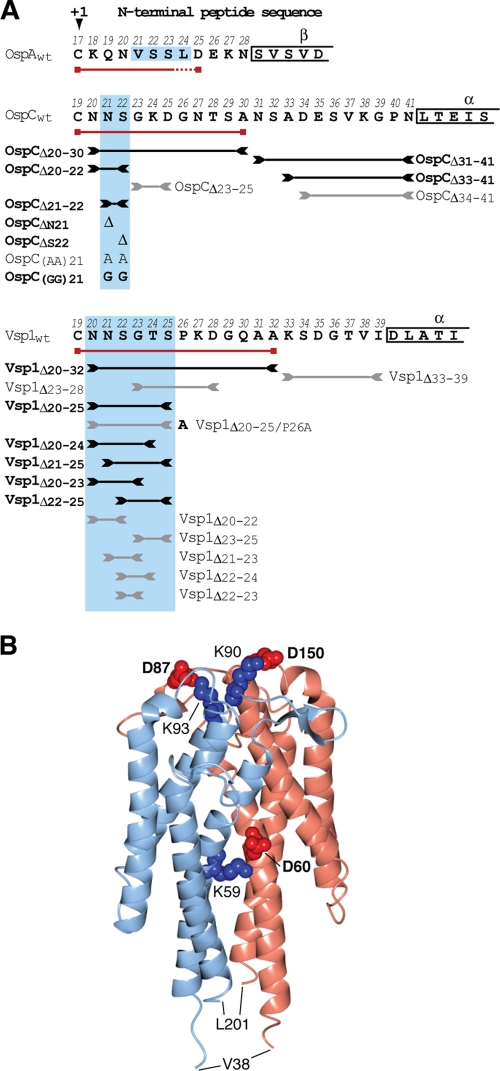
Fig. 1.
Genotypes and phenotypes of OspC and Vsp1 mutants. (A) N-terminal sequences of mature lipoproteins OspA, OspC, and Vsp1 are shown in single-letter amino acid code. The +1 position Cys residue is marked with an arrowhead. Numbers above the residues indicate their positions in the prolipoprotein, including the cleaved signal peptide. Greek letters above boxed residues indicate secondary structure elements as determined by X-ray crystallography (19, 30, 32, 34). Red lines with boxed ends underline the minimum tether sequences required for surface localization of mRFPΔ4. Lines flanked by inverted arrowheads span the peptides deleted in the respective tether mutants. Black lines/bold letters mark mutants with non-wild-type phenotypes. Gray lines/regular letters mark mutants with a wild-type phenotype. Boxes shaded in light blue indicate the essential tether motifs of OspA, OspC, and Vsp1. Mutant nomenclature follows that of earlier publications (58, 59). Briefly, modified residues are numbered according to their prolipoprotein positions; numbers in lipoprotein tether-reporter fusions indicate the C-terminal tether residues present in the fusions. (B) A ribbon representation of the Vsp1 tertiary structure (Protein Data Bank ID 2GA0) (32) was generated using the CCP4 software for Macintosh (version 2.4.3) (50). The two Vsp1 chains are colored light blue and pink. Residues involved in salt bridging of the monomers are highlighted in red (Asp) or blue (Lys) spheres representing the Cα and side chain atoms. The bolded residues were mutated to yield the Vsp1 monomer. Val38 and Leu201 are the first and last residues visible in the crystal structure.