Abstract
Cells carrying the thermosensitive nrdA101 allele are able to replicate entire chromosomes at 42°C when new DNA initiation events are inhibited. We investigated the role of the recombination enzymes on the progression of the DNA replication forks in the nrdA101 mutant at 42°C in the presence of rifampin. Using pulsed-field gel electrophoresis (PFGE), we demonstrated that the replication forks stalled and reversed during the replication progression under this restrictive condition. DNA labeling and flow cytometry experiments supported this finding as the deleterious effects found in the RecB-deficient background were suppressed specifically by the absence of RuvABC; however, this did not occur in a RecG-deficient background. Furthermore, we show that the RecA protein is absolutely required for DNA replication in the nrdA101 mutant at restrictive temperature when the replication forks are reversed. The detrimental effect of the recA deletion is not related to the chromosomal degradation caused by the absence of RecA. The inhibition of DNA replication observed in the nrdA101 recA mutant at 42°C in the presence of rifampin was reverted by the presence of the wild-type RecA protein expressed ectopically but only partially suppressed by the RecA protein with an S25P mutation [RecA(S25P)], deficient in the rescue of the stalled replication forks. We propose that RecA is required to maintain the integrity of the reversed forks in the nrdA101 mutant under certain restrictive conditions, supporting the relationship between DNA replication and recombination enzymes through the stabilization and repair of the stalled replication forks.
INTRODUCTION
Ribonucleotide reductase (RNR) is the only enzyme specifically required for the formation of deoxyribonucleotides (deoxynucleoside triphosphates[dNTP]), the precursors of DNA synthesis in Escherichia coli. RNR is a 1:1 complex of two nonidentical subunits, called R1 and R2, encoded by the genes nrdA and nrdB, respectively (8). The best-known defective RNR mutant in E. coli contains a thermolabile R1 subunit, coded by the nrdA101 allele. This allele carries a missense mutation, causing a change in amino acid 89 (L89P) (29). The activity of the enzyme measured in crude extracts of nrdA101 strains is limited to 6% of the wild type at 25°C (9), and the dNTP pool is lower than that of wild type even at the permissive temperature of 30°C (24). The RNR101 protein is inactivated in vitro after 2 min at 42°C (9) although a thermoresistant period of 50 min has been observed in vivo in the nrdA101 mutant strain (17). This thermoresistant period of DNA synthesis has been ascribed to the protection of RNR101 by the replication hyperstructure (15–17, 30, 31) and is extended when RNA and/or protein synthesis is inhibited, allowing cells to replicate entire chromosomes at 42°C (17). This feature has been related to a decrease in the overlap of replication rounds after the inhibition of new DNA initiation events (I. Salguero, E. López-Acedo, and E. C. Guzmán, submitted for publication).
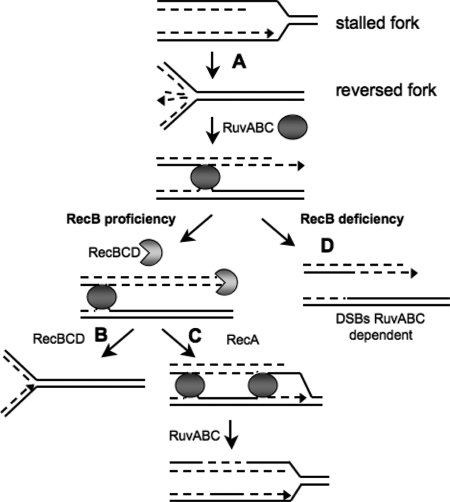
In a previous work we showed that in the nrdA101 mutant strain at 30°C, the DNA replication progresses with the help of replication fork reversal (RFR) (15). This mechanism has been extensively studied by Michel and coworkers (25). In this model, newly synthesized strands anneal, forming a Holliday junction (HJ) (Fig. 1A). In a rec-proficient background, this intermediate can be processed without generating DNA double-strand breaks (DSBs) by the action of RecBCD exonuclease that degrades the DNA tail (Fig. 1B) or by using the recombination proteins RecBCD and RecA and the HJ-specific resolvase RuvABC (Fig. 1C). Nevertheless, in the absence of RecBCD activity (Fig. 1D), resolution of the HJ generated by RFR is performed by the RuvABC resolvase and leads to DNA breakage, so these DSBs are dependent on RuvABC activity in a recB-deficient background. According to the RFR model, the occurrence of this process at the stalled forks generated under restrictive conditions can be verified by testing whether there is an increase of DSBs in a recB-deficient background and determining whether these DSBs are dependent on RuvABC resolvase activity (Fig. 1 D) (25).
Fig. 1.
Fate of the stalled forks. In the first step (A), the replication fork is arrested, causing fork reversal and forming an HJ. In recombination-proficient cells (B), RecBCD initiates RecA-dependent homologous recombination, and RuvABC resolves the resulting double HJ. In the absence of RecBCD (C), resolution of the HJ by RuvABC leads to DSBs at the stalled replication fork. Solid line, parental chromosome; dashed line, newly synthesized strand; solid disk, RuvAB; incised disk, RecBCD.
Apart from the requirement of RecA to resolve reverted replication forks by the homologous recombination pathway, RecA binds to single-strand DNA regions generated by replication, maintaining structural integrity of the fork when it is impeded from progressing normally (34). According to this finding, it has also been proposed that RecA plays an important role protecting the replication forks when they stall due to encountering lesions on the DNA molecule (4, 5, 18).
In this work we have studied the role of the recombination enzymes on the progression of the DNA replication forks in the nrdA101 mutant at 42°C in the presence of rifampin. We have found that the nrdA101 mutant completes chromosomal replication with the help of RFR and the assistance of RecA protein. We propose that RecA is absolutely required for stabilizing the reversed forks in addition to its recombination role and, hence, for DNA replication under these restrictive conditions. This requirement would support the relationship between DNA replication and RecA by elucidating a role for RecA in maintaining a successful DNA structure at the replication forks under stress conditions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli JS1018 (nrdA101 thyA arg his thi malA rpsL su xyl mtl) is a Pol+ Thy− low-requirement derivative from the E1011 strain obtained from R. McMacken (Stanford University, Stanford, CA). All strains used in this work were JS1018 derivatives (Table 1). Strains were constructed by the standard P1 transduction or Hfr conjugation method, selecting for the appropriate resistance. Given that the priA null mutant may accumulate suppressor mutations, the nrdA101 priA2::Kam double mutant (JS891) was constructed and kept in the presence of the pAM-priA+ plasmid, which carries a spectinomycin (Spec) resistance gene, the wild-type priA gene, and an isopropyl-β-d-thiogalactoside (IPTG)-dependent replication origin. This plasmid allows cell growth in a priA+ context if IPTG and Spec are present and also the priA+ gene, which was lost in the absence of IPTG and Spec for the experiments (14). Finally, all the recombination mutants as well as the priA2::Kam strain were checked for the characteristic UV sensitivity.
Table 1.
Strains used in this work
| Strain | Relevant genotype | Origin or construction |
|---|---|---|
| JS1018 | nrdA101 thyA arg his thi malA Lr rpsL mtl xyl | This lab |
| JS627 | JS1018 Δ(recA-srl)::Tn10 | P1 transduction of Δ(recA-srl)::Tn10 in JS1018 |
| JS628 | JS1018 recB258::Tn10 | P1 transduction of recB258::Tn10 in JS1018 |
| JS704 | JS1018 ΔruvABC::Cm | P1 transduction of ΔruvABC::Cm in JS1018 |
| JS705 | JS1018 ΔruvABC::Cm recB258::Tn10 | P1 transduction of recB258::Tn10 in JS704 |
| JS767 | JS1018 recG263::Kan | P1 transduction of recG263::Kan in JS1018 |
| JS768 | JS1018 recG263::Kan recB258::Tn10 | P1 transduction of recB258::Tn10 in JS767 |
| JS769 | JS1018 ΔrecBC::Ap Δ(recA-srl)::Tn10 | Hfr conjugation of ΔrecBC::Ap Δ(recA-srl)::Tn10 in JS1018 |
| JS868 | JS1018 pAM-priA+ | Tranformation of JS1018 with pAM-priA+ |
| JS881 | JS1018 Δ(recA-srl)::Tn10/pRecA | Transformation of JS627 with pRecA |
| JS882 | JS1018 Δ(recA-srl)::Tn10/pRecAS25P | Transformation of JS627 with pRecAS25P |
| JS891 | JS1018 priA2::Kam/pAM-priA | P1 transduction of priA2::Kam in JS1018/pAM-priA |
| JK607 | JS1018 nrdA+ yfaL::Tn5 | P1 transduction of nrdA+ yfaL::Tn5 in JS1018 |
| JK625 | JK607 Δ(recA-srl)::Tn10 | P1 transduction of Δ(recA-srl)::Tn10 in JK607 |
| JK626 | JK607 recB258::Tn10 | P1 transduction of recB258::Tn10 in JK607 |
| JK706 | JK607 ΔruvABC::Cm | P1 transduction of ΔruvABC::Cm in JK607 |
| JK707 | JK607 ΔruvABC::Cm recB258::Tn10 | P1 transduction of recB258::Tn10 in JK706 |
| JJC275 | Δ(recA-srl)::Tn10 (mini-F recA+ Ampr) | B. Michel |
| JJC777 | recB258:: Tn10/pDWS2 (pBR322 recBCD+) | B. Michel |
| JJC754 | ΔruvABC::Cm | B. Michel |
| JJC1130 | Hfr PK191 rpsL ΔrecBC::Ap Δ(recA-srl)::Tn10/pDWS2 | B. Michel |
| JJC1398 | priA2::Kam sfiA11/pAM-priA | B. Michel |
| JJR750 | JK607 sulA::Mud Apr | This lab |
| JJR751 | JS1018 sulA::Mud Apr | This lab |
| N3793 | recG263::Kan | CGSC 7552 |
Bacteria were grown by shaking at 30°C in M9 minimal medium (MM9) containing M9 salts, 2 μg/ml thiamine, 0.4% glucose, 20 μg/ml of the required amino acids, 5 μg/ml thymidine, and 0.2% Casamino Acids, after a 1:200 dilution of an overnight culture. Growth was monitored by light absorbance at 550 nm. Incubation at 42°C or the addition of rifampin (150 μg/ml) was initiated at an optical density at 550 nm of 0.1. The cultures from the nrdA101 priA2::Kam strain were obtained as previously described in the literature (13, 14). Briefly, cells containing pAM-priA were grown overnight in Luria broth (LB) in the presence of IPTG and Spec. Then, the culture was diluted 1,000-fold in minimal medium without IPTG or Spec and grown again overnight. This first minimal medium culture was diluted one more time 200-fold in minimal medium with no IPTG or Spec and then grown until an optical density at 550 nm (OD550) of 0.1. At this point, cells were shifted at 42°C in the presence of rifampin. A sample of the culture was taken before the addition of the rifampin, and dilutions were plated in nutrient agar. The Spec sensitivity of the grown colonies was verified, indicating that the cells used for the experiment did not have the plasmid and therefore were priA mutants.
Measure of DSBs by PFGE.
Measurements of DSBs were performed as described previously in the literature (26, 35). Briefly, for chromosome labeling, cells were grown in minimal medium with 0.2% Casamino Acids in the presence of 5 μCi/ml of [methyl-3H]thymidine (20 Ci/mmol) (ICN) until the culture reached an OD550 of 0.1. Cells were collected, washed, and embedded in agarose plugs. Gentle lysis was performed in the plugs before they were used for pulsed-field gel electrophoresis (PFGE). The proportion of migrating DNA was determined by cutting each lane into slices and counting the tritium present in the wells and in the gel slices. The linear DNA values were expressed as the ratio (percent) between counts per minute (cpm) in the gel slices and the cpm in the gel slices plus well.
DNA synthesis measurements.
DNA synthesis was determined by growing the cells in MM9 medium containing 1 mCi/ml of [methyl-3H] thymidine (20 Ci/mmol) (ICN) and assaying the radioactive acid-insoluble material.
Flow cytometry.
DNA content per cell was measured by flow cytometry using a Bryte HS (Bio-Rad) flow cytometer, as previously described (12, 38). When the cultures reached an OD550 of 0.1, a portion was transferred into another flask, and rifampin (150 μg/ml) and cephalexin (50 μg/ml) were added to inhibit new rounds of chromosome replication and cell division, respectively. These treated cultures were grown for an additional 4 h with continuous shaking, after which 400 ml of each culture was added to 7 ml of 74% ethanol. Approximately 1.5 ml of each fixed sample was centrifuged, and the pellets were washed in 1 ml of ice-cold staining buffer (10 mM Tris, 10 mM MgCl2, pH 7.4, in sterile distilled H2O [dH2O]) and resuspended in 65 μl of staining buffer and 65 μl of staining solution (40 μg/ml ethidium bromide and 200 μg/ml mithramycin A). Cells were incubated on ice in the dark for at least 30 min and analyzed on a Bryte-HS Flow Cytometer (Bio-Rad) at 390 to 440 nm.
RESULTS
The nrdA101 mutant undergoes replication fork reversal in the presence of rifampin.
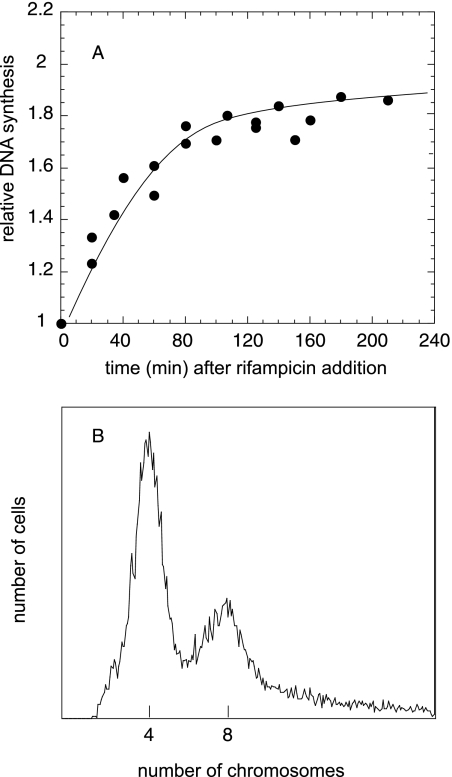
It has been shown that the nrdA101 mutant is able to complete chromosome replication at the restrictive temperature if initiation of replication is repressed by protein synthesis inhibition (17; also Salguero et al., submitted). The complete replication of the chromosome at 42°C is unexpected as the protection of RNR101 by the replication hyperstructure does not cover an entire replication cycle in exponentially growing cultures incubated at 42°C (17).
We analyzed the progression of the replication forks in the nrdA101 mutant after the addition of rifampin at 42°C. To do this, first we determined whether the progression of the replication forks proceeded without stalling. Arrest of replication forks is known to cause DSBs (19), and quantification of this parameter indicates the level of stalled replication forks caused by the treatment (2, 22, 23). We measured the amount of DSBs by using pulsed-field gel electrophoresis combined with cell lysis in agarose plugs. To detect the DSBs, it is necessary to prevent degradation of linear DNA and repair of broken DNA. This can be achieved by inactivating RecBCD activity using a recB-deficient background although recA recD doubly deficient strains can also be used (26).
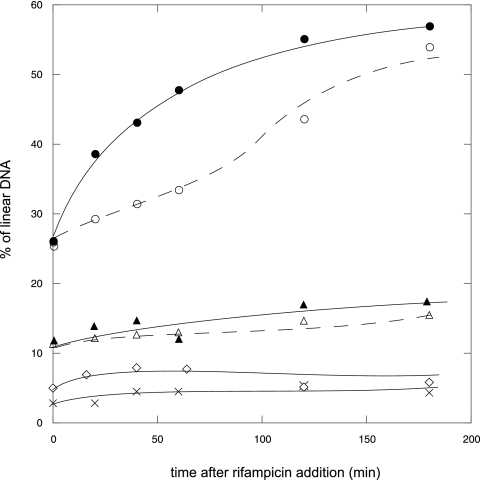
DSBs in the nrdA101 recB mutant after the addition of rifampin at 30°C and at 42°C were quantified (Fig. 2), and results indicated that the number of DSBs in this strain increases at both 30°C and 42°C, suggesting an increment in the number of stalled forks during the progression of replication. Given that the DNA replication in nrdA101 mutants is completed after addition of rifampin at 30°C and at 42°C, these stalled replication forks must be restarted in a recombination-proficient strain.
Fig. 2.
Percentage of linear DNA in nrdA+ recB, nrdA+ recB ruvABC, nrdA101 recB, and nrdA101 recB ruvABC strains after incubation at 30°C or 42°C in the presence of rifampin. Strains are as follows: nrdA101 recB strain at 30°C (○)or 42°C (●), nrdA+ recB strain at 30°C (▵) or 42°C (▴), nrdA101 recB ruvABC strain at 30°C or 42°C (♢), and nrdA+ recB ruvABC strain at 30°C or 42°C (×) in the presence of rifampin. The represented values are the averages of two independent experiments.
In a previous work, we have demonstrated that the nrdA101 mutant undergoes RFR to restart the stalled replication forks (15). This suggested to us that RFR could be used to reactivate the DNA replication forks at 42°C in the presence of rifampin. According to the RFR model, the formation of DSBs would result from the action of the RuvABC resolvase. To establish the origin of the DSBs, the level of DSBs was estimated in an nrdA101 recB ruvABC strain. We found that these levels were invariable and markedly lower than in the respective nrdA101 recB ruv+ counterpart strains (Fig. 2). As RuvABC is a specific resolvase for HJs, these results indicate the occurrence of replication fork reversal in the nrdA101 mutant after the addition of rifampin at both temperatures. Because RFR is one of the mechanisms known to restart the stalled replication forks, we could infer that in the nrdA101 strain under the addition of rifampin at 42°C the progression of the replication forks is disturbed but proceeds along the chromosome by the occurrence of RFR.
Quantification of the amount of linear DNA in the nrdA+ recB strain showed no substantial increase in the number of DSBs during the progression of the replication cycle after rifampin addition, indicating that RFR is associated with the nrdA101-defective background. It should be noted that, according to our previous results, the level of DSBs in the nrdA101 recB strain in exponential growth (i.e., before the addition of rifampin) is higher than that observed in the nrdA+ recB strain, as a defective RNR101 impairs the replication progression even at permissive temperatures and replication forks stop more frequently (15).
Role of recombination enzymes in the DNA replication of the nrdA101 mutant in the presence of rifampin.
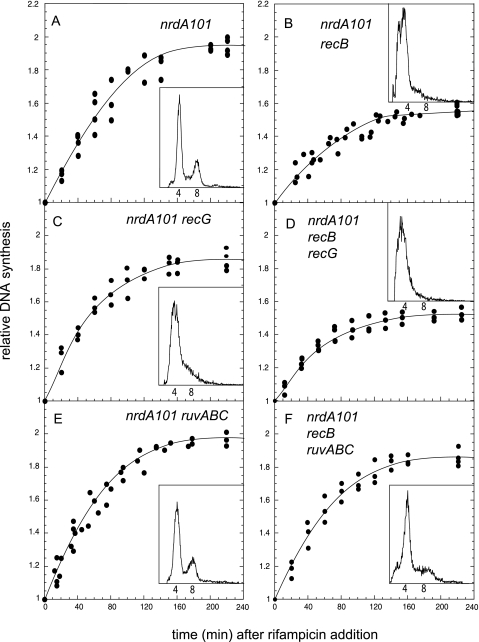
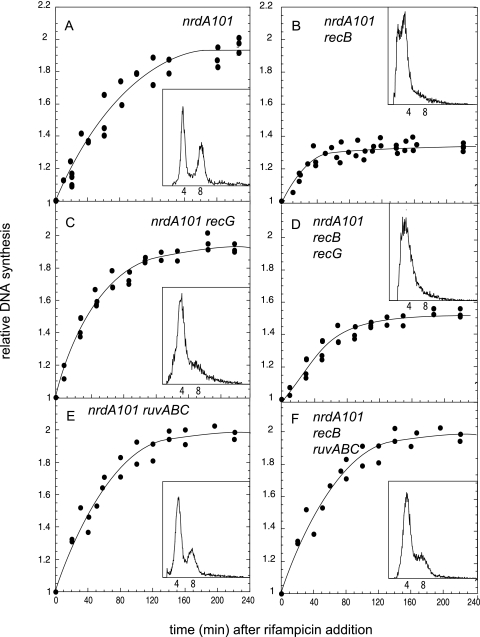
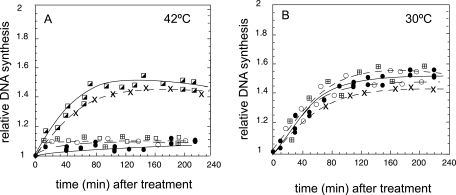
To further verify the occurrence of RFR in the nrdA101 mutant under this restrictive condition, we determined whether the deficiency of the recombination enzymes could affect DNA synthesis at 42°C in the presence of rifampin. We quantified the runout of the DNA replication (i.e., the amount of DNA synthesized after inhibition of the initiation of replication) by determining the [methyl-3H]thymidine incorporation on the DNA at 30°C (Fig. 3) and at 42°C (Fig. 4) and the flow cytometry profiles of the different nrdA101 strains after 4 h of rifampin and cephalexin addition (Fig. 3 and 4, insets). We analyzed an nrdA101 mutant that was also deficient for RecB, RuvABC, or RecG activity and the nrdA101 recB ruvABC and nrdA101 recB recG triple mutants. The results showed that the nrdA101 recB mutant was partially affected in its capacity to replicate DNA at either 30°C (Fig. 3B) or 42°C (Fig. 4B) in the presence of rifampin. The flow cytometry profile after 4 h of treatment showed a broad distribution without any discrete peaks corresponding to four or eight chromosomes per cell, as observed in the nrdA101 strain after the shift in the presence of rifampin (to inhibit DNA initiation) and cephalexin (to inhibit cell division) (Fig. 3A and 4A). This indicates that, in the absence of the RecB, replication in the nrdA101 mutant stops stochastically after a thermoresistant period without completing the rounds of DNA replication. Regarding the deficiency of RecG and RuvABC, the runout results shown in Fig. 4C and E, respectively, indicate that the deficiency of these proteins did not alter the chromosomal replication. This suggests that, in the presence of an active RecB, functional RecG and/or RuvABC is not required or that they both can be used redundantly when the nrdA101 strain replicates DNA at 42°C in the presence of rifampin. The flow cytometry profile showed no alterations when RuvABC was absent although segregation of the chromosomes must be partially affected in the nrdA101 recG mutant, as the peak corresponding to cell with eight chromosomes was not completely resolved under these experimental conditions. Similar results were found 4 h after the addition of rifampin and cephalexin at 30°C, suggesting that this effect corresponds to a feature related to RecG deficiency (Fig. 3C and E).
Fig. 3.
Runout of DNA synthesis and flow cytometry profile (insets) of the indicated strains after the addition of rifampin at 30°C.
Fig. 4.
Runout of DNA synthesis and flow cytometry profile (insets) of the indicated strains after the addition of rifampin at 42°C.
Consistent with the occurrence of RFR in the nrdA101 mutant after the addition of rifampin at both 42°C and 30°C, we found that RuvABC deficiency but not RecG absence suppressed the alterations of DNA synthesis due to the lack of RecB, allowing complete DNA chromosomal replication and segregation in the nrdA101 recB ruvABC mutant (Fig. 3D and F, respectively, and 4D and F). This would suggest that, in the absence of RecB and RuvABC, the reversed replication fork could be regressed to the initial position with no detrimental effect.
PriA protein is not required for DNA replication of an nrdA101 mutant to complete chromosomal replication under restrictive conditions.
Once the stalled replication forks have been processed and if the replication machinery is removed from the chromosome after the stop, restarting replication would require the assembly of a new replisome at the point where replication was disrupted. PriA triggers this step by allowing the assembly of a new replication complex at a site different from oriC (10, 20, 33).
Given that replication forks stall and reverse during DNA replication in the presence of rifampin at 42°C in the nrdA101 mutant, we studied the potential role of PriA in restarting replication under these conditions. To achieve this, we constructed a double nrdA101 priA mutant to determine whether the defective nrdA101 strain was able to complete chromosome replication in the presence of rifampin at the restrictive temperature in the absence of PriA. The priA mutant acquires suppressor mutations very easily (32); for this reason, the experiments with the double nrdA101 priA mutant were performed as described in the Materials and Methods section.
To verify the completion of the chromosomal replication in the nrdA101 priA strain, DNA synthesis and flow cytometry analysis were carried out on cultures treated with rifampin and cephalexin at 42°C (Fig. 5). These results show that the nrdA101 priA double mutant was able to finish the replication under this restrictive condition, indicating that PriA is dispensable for the replication of the nrdA101 strain in the presence of rifampin at 42°C and suggesting that the replisome was not disassembled by the reversion of the replication forks.
Fig. 5.
Runout of DNA synthesis (A) and flow cytometry profile (B) of the nrdA101 priA mutant after rifampin addition at 42°C.
RecA protein is absolutely required for DNA replication in the nrdA101 mutant under restrictive conditions.
Regarding the ability of recombination mutants to synthesize DNA at 42°C in the presence of rifampin, the nrdA101 recA strain showed a surprising result among all the recombination-defective nrdA101 mutants. Figure 6 and Table 2 show that the nrdA101 mutant demonstrates an absolute requirement for RecA to replicate the chromosome at 42°C in the presence of rifampin, although residual DNA synthesis was observed in the absence of the drug under restrictive conditions (Fig. 6A) or in the presence of rifampin at 30°C (Fig. 6B). Nevertheless, the DNA synthesis of the nrdA101 recA strain after inhibition of new initiations at 30°C is partially affected compared to its recA+ counterpart (Table 2), and the completion of chromosome replication was not observed (data not shown). This effect has been previously reported in different recA-deficient backgrounds (37).
Fig. 6.
(A) Runout of DNA synthesis of the nrdA101 recA strain at 42°C (◪) or after rifampin addition (●), chloramphenicol addition (⊞), and amino acid starvation (○) at 42°C; of the nrdA101 recA recB strain after rifampin addition at 42°C (□); and of the nrdA+ recA strain after rifampin addition at 42°C (×). (B) Runout of DNA synthesis of the nrdA101 recA strain after rifampin addition (●), chloramphenicol addition (⊞), and amino acid starvation (○) at 30°C and of the nrdA+ recA strain after the addition of rifampin at 30°C (×).
Table 2.
Relative increment in the amount of DNA in nrdA+ and nrdA101 strains and recA derivatives after 4 h of rifampin addition at 30°C or 42°C and after incubation at 42°C in the absence of the drug
| Strain genotype | Relative increase DNA (%) under the indicated conditiona |
||
|---|---|---|---|
| + Rif at 30°C | + Rif at 42°C | − Rif at 42°C | |
| nrdA+ | 52 | 55 | |
| nrdA+recA | 36 | 47 | |
| nrdA101 | 90 | 90 | 48 |
| nrdA101 recA | 41 | 5 | 45 |
Values are based on ΔG. The presence (+ Rif) and absence (− Rif) of rifampin is indicated.
We showed that the sudden stop of the DNA synthesis in the nrdA101 recA mutant at 42°C was not associated with a specific effect of the rifampin in RNA synthesis inhibition, as similar results were obtained in the presence of chloramphenicol or under amino acid starvation (Fig. 6A). Finally, we showed that this effect was caused by the absence of RecA in the nrdA101 mutant as DNA synthesis in the nrdA+ recA strain was not inhibited under similar conditions (Fig. 6A and Table 2).
RecA protein is essential for SOS induction. Therefore, it could be possible that DNA replication in an nrdA101 mutant at 42°C in the presence of rifampin required some SOS protein, induced prior to the abrogation of protein synthesis. If this was the case, constitutive SOS induction should be observed in the nrdA101 mutant at 30°C. We tested a potential constitutively induced SOS response provoked by the nrdA101 allele by using a sulA::lacZ fusion in the nrdA101 strain background, in which the level of expression of the lacZ gene from the sulA promoter is a measure of SOS induction. No SOS induction was found in the nrdA101 mutant compared to SOS levels in its nrdA+ counterpart (207 and 225 Miller units, respectively) although an important increase was observed after UV irradiation in both strains (670 and 640 Miller units, respectively). Therefore, involvement of the SOS response could be ruled out in the replication of the nrdA101 mutant at 42°C in the presence of rifampin.
A second possibility could be that the lack of recombination processes was related to the phenotype caused by the absence of RecA. However, this seems unlikely because the nrdA101 recBCD mutant does not show such abrupt arrest of DNA synthesis (Fig. 4B), and, according to the RFR model, once the fork is reversed, the next step where RecA could have a role is in the processing of the HJ when single-stranded DNA is generated by the RecBCD exonuclease, which is subsequently coated by RecA (Fig. 1C). Therefore, the lack of the RecA function in recombination would not have any impact in a recB-deficient background. To rule out this hypothesis, we determined the runout of the nrdA101 recA recB triple mutant at 42°C in the presence of rifampin. Figure 6A shows that DNA synthesis was not restored by abrogating RecBCD activity, suggesting that RecA deficiency could be altering the replication fork integrity in addition to abolishing recombination-repair processes at the replication point.
DNA degradation cannot explain the absence of newly synthesized DNA in the nrdA101 recA mutant under restrictive conditions.
It is well documented that DNA degradation is observed in recA mutants. Exponentially growing recA mutant cells display pronounced DNA degradation, which starts at the sites of DNA damage and depends on RecBCD nuclease (ExoV) activity (23, 28). To find out whether this feature corresponds with the incapacity to replicate chromosomal DNA, we analyzed DNA degradation at 30°C and 42°C in the presence of rifampin.
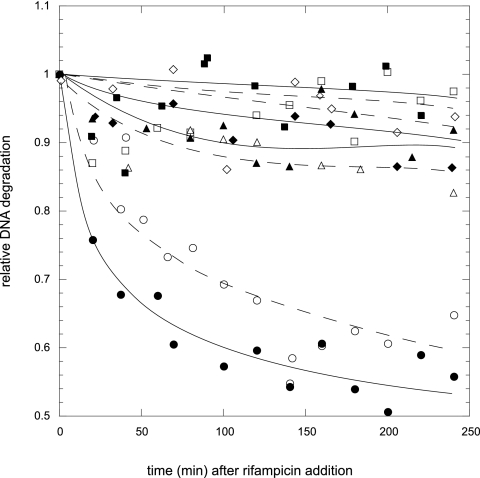
Cultures of the nrdA101 and nrdA101 recA strains were labeled with [methyl-3H]thymidine. By the time the cultures reached an OD550 of 0.1, one portion of the culture was filtered and resuspended in the same medium free of [methyl-3H]thymidine, treated with rifampin, and grown at either 30°C or 42°C, and samples were taken at different time points. As shown in Fig. 7, DNA degradation was observed in the nrdA101 recA strain in the presence of rifampin at both temperatures to a much higher extent than in the rec+ isogenic strain. To verify that this DNA degradation was due to the RecA protein deficiency, we transformed the nrdA101 recA strain with the plasmid pRecA, a pT7-7 derivative carrying the recA gene (36). As expected, the stability of the DNA in the nrdA101 recA strain was restored by the presence of RecA (Fig. 7). Furthermore, we found that DNA degradation was completely suppressed in the nrdA101 recA recB strain (Fig. 7). Given that DNA synthesis in the nrdA101 recA recB strain was inhibited under restrictive conditions, this indicates that the abrupt stop of DNA synthesis in the nrdA101 recA mutant is not related to the DNA degradation carried out by RecBCD.
Fig. 7.
DNA degradation in the nrdA101 (□ and ■), nrdA101 recA (○ and ●), nrdA101 recA recB (▵ and ▴), and nrdA101 recA/pRecA (♢ and ♦) strains after the addition of rifampin at 30°C (open symbols) and 42°C (closed symbols).
Recombination activity of the RecA protein only partially recovers DNA synthesis in an nrdA101 mutant at 42°C.
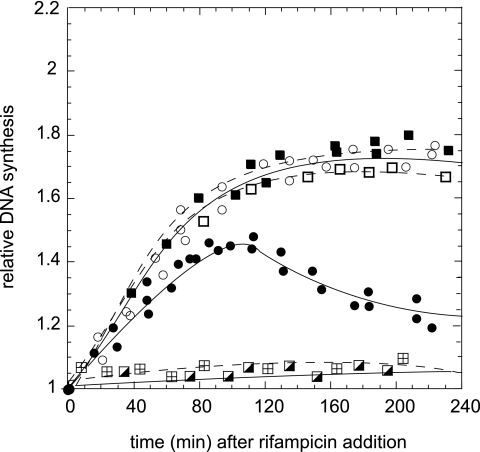
The results described above indicate that DNA degradation is suppressed by the presence of the wild-type RecA protein. Next, we investigated whether the DNA synthesis inhibition in the presence of rifampin at 42°C in the nrdA101 recA strain was also rescued by the functioning RecA protein from the plasmid pRecA. The runout experiments showed that DNA replication of the nrdA101 recA/pRecA strain after the addition of rifampin at 42°C was restored (Fig. 8), indicating that RecA deficiency was responsible for the arrest of DNA synthesis observed under restrictive conditions in the nrdA101 recA mutant.
Fig. 8.
Runout of DNA synthesis of the nrdA101 recA (⊞), nrdA101 recA/pRecA (○), nrdA101 recA/pRecA(S25P) (●), and nrdA101 recA recB (◪) strains after the addition of rifampin at 42°C and of the nrdA101 recA/pRecA (■) and nrdA101 recA/pRecA(S25P) (□) strains after the addition of rifampin at 30°C.
RecA is involved in the homologous recombination processes; however, the results presented above suggest that the primary role of RecA on the DNA replication of the nrdA101 mutant at 42°C in the presence of rifampin is not associated with the recombination process itself. Apart from its function in recombination, RecA has a role in protecting and rescuing stalled or collapsed replication forks (3, 5). To study whether the deficiency of the role of RecA in maintaining the replication fork progression was related to the inhibition of DNA replication observed in the nrdA101 recA mutant at 42°C in the presence of rifampin, we used a RecA protein with the mutation S25P [RecA(S25P)]. This altered protein is able to carry out DNA repair, homologous recombination, and SOS induction but fails in recovering the alterations of replication progression in DNA polymerase mutants (36). Runout DNA synthesis of the nrdA101 recA/pRecA(S25P) strain at 30°C and 42°C in the presence of rifampin was performed. The results indicated that RecA(S25P) suppressed the deficiency of DNA synthesis in the presence of rifampin at 30°C but only partially suppressed the RecA immediate stop of DNA replication of the nrdA101 recA mutant in the presence of rifampin at 42°C (Fig. 8). These results indicate that DNA synthesis in an nrdA101 recA mutant after the addition of rifampin at 42°C is only partially recovered by a RecA form fully active for recombination functions but defective in stabilizing stalled replication forks. Furthermore, DNA degradation is observed by the time the replicating forks stalled. Crucially, it is the activity of RecA in protecting and rescuing the replication forks, absent in the RecA(S25P) protein, that is required for the nrdA101 mutant to replicate and maintain the integrity of the entire chromosome in the presence of rifampin at the nonpermissive temperature. Moreover, this role of RecA seems to be dispensable for replication progression under permissive conditions.
DISCUSSION
The nrdA101 mutant, is able to replicate the entire chromosome at 42°C when initiation of replication is inhibited, i.e., after rifampin or chloramphenicol addition or by amino acid starvation. We show here that the DNA replication progression of the nrdA101 mutant under this restrictive condition requires the principal recombination enzymes in two different processes: one of them related to the processing of the stalled replication forks and the other one related to the protection of the stalled replication forks.
Recombination enzymes involved in the processing of stalled forks help the nrdA101 mutant to complete chromosome replication at the restrictive temperature after rifampin addition.
Completion of chromosomal replication of the nrdA101 mutant at 42°C in the presence of rifampin indicates that RNR101 must be active to provide the required dNTPs for DNA synthesis. The RNR101 protection from thermal inactivation by the replication hyperstructure has been proposed to explain the residual DNA synthesis of the nrdA101 mutant after the temperature shift to 42°C (15–17, 31). Nevertheless, this residual DNA synthesis is not enough to yield complete replicated chromosomes, as observed after rifampin addition under restrictive conditions. In a recent work we have proposed that the inhibition of new initiation events following the addition of rifampin would improve the progression of the ongoing replication forks by lowering the number of replication forks per chromosome (Salguero et al., submitted). Here, we have examined the progression of the replication forks under these conditions.
DNA replication is not a continuous process because replication forks stall and need to be rescued by the action of recombination enzymes (21, 25). The results presented here show that in the nrdA101 mutant, the replication forks stalled and reversed to achieve the end of the replication round both at 30°C and at 42°C in the presence of rifampin. According to the RFR model, the occurrence of RuvABC-dependent DSBs in a recB-deficient background would be an indicator of the amount of stalled replication forks that have reverted. We showed that the nrdA101 recB mutant, but not the nrdA+ recB strain, exhibits an increase in DSBs after the addition of rifampin at 30°C and 42°C. We found that, in the nrdA101 recB strain, the induced DSBs are dependent on RuvABC activity, indicating that they are generated after resolution of an HJ created by the reversion of the stalled forks. Therefore, this indicates that there is an increase in the number of stalled replication forks in the nrdA101 mutant that would proceed with the help of RFR in a rec+ background. We show that the RecB deficiency impairs the progression of the replication forks of the nrdA101 mutant at 30°C and 42°C in the presence of rifampin as the runouts of DNA synthesis in the nrdA101 recB double mutant were less than half of those observed in an nrdA101 strain, most likely due to the unrepaired DSBs generated at stalled replication forks. Moreover, this detrimental effect was recovered specifically by RuvABC deficiency but not by RecG absence, supporting the observation that the DSBs observed by PFGE were RuvABC dependent.
The occurrence of RFR at the replication forks has been observed in several replication mutants affected in DNA elongation, but RFR does not occur in mutants impaired in replication initiation or in replicative topoisomerases (27), which implies that forks arrested by different means differ in their structure or in the presence of different sets of associated replication proteins. A good example of this is the arrest of replication forks by deoxynucleotide starvation. We have shown that the RFR process is induced when DNA synthesis is inhibited by hydroxyurea addition. However, a significant increase in the RFR level is not observed when replication forks are arrested in the RNR-defective nrdA101 mutant at a restrictive temperature, compared to that observed at 30°C (16). Our results here show that, in the presence of rifampin, the stalled replication forks in the nrdA101 mutant became predisposed to reverse at 42°C, in contrast with what happens when the drug is not added at the restrictive temperature. At least two features concerning the inhibition of RNA and/or protein synthesis could be related to the structure of the stalled replication forks: the first one is the decrease in the number of replication forks running on the bacterial chromosome, and the second is the absence of RNA transcripts. Both conditions could be associated with changes in DNA topology (6, 7, 39), and, consequently, they could be related to the capacity of the stalled replication forks to be reversed. Given that complete chromosomal replication in the nrdA101 mutant at the restrictive temperature is possible in the presence of rifampin but not in its absence, this suggests that the reversion of the replication forks would contribute to improved DNA replication progression under certain restrictive conditions.
In addition to the reversion, stalled forks must be restarted. The key protein engaged in this process is PriA, which recruits the replication proteins to the fork (10). We found that the reversed replication forks generated under the addition of rifampin at 42°C in the nrdA101 mutant do not require PriA to complete chromosomal replication. This result was expected according to the notion of the structural protection of the RNR101 by the replication hyperstructure (17). The requirement of PriA would imply that the replication hyperstructure would disassemble from the stalled forks and that the unprotected RNR101 would be inactivated before replication was completed. Our results support the idea that, in the presence of rifampin at 42°C, the replication hyperstructure must be stable throughout chromosomal replication, maintaining the thermosensitive RNR101 as functionally active to reach the end of the chromosome.
The RecA protein is absolutely required for the progression of replication forks.
We found that the RecA deficiency provoked a dramatic effect on DNA synthesis in the nrdA101 mutant when it was incubated at 42°C under inhibition of initiation. Furthermore, we showed that this cessation of DNA replication is not related to a defect in SOS induction, is not associated with the absence of recombination events, and is not the consequence of the extensive DNA degradation associated with RecA deficiency.
Apart from the aforementioned functions, RecA has a role in protecting and rescuing stalled or collapsed replication forks (3, 5). In this sense, it has been proposed that RecA is required to maintain the integrity of the arrested replication forks until they can resume DNA replication once the lesion has been removed (5).
We studied whether the inhibition of DNA replication observed in the nrdA101 recA mutant at 42°C in the presence of rifampin was related to the role of RecA in maintaining the stalled replication forks. This function is lost in the mutant protein RecA(S25P), although the mutant is able to carry out DNA repair, homologous recombination, and SOS induction (36). The ectopically expressed RecA(S25P) protein fully reestablished the DNA runout at 30°C but only partially restored DNA synthesis under the restrictive condition for 80 to 90 min (up to 40%), and this was followed by extensive DNA degradation. This reflects that RecA(S25P) might be defective in protecting or maintaining the replication forks generated at 42°C in the presence of rifampin in such a way that RecBCD would eventually degrade them. Altogether, these observations point to an exceptional role of RecA under these circumstances, different from processing the reverted replication forks by homologous recombination but related to the stabilization of these stalled forks under restrictive conditions.
Regarding the specific requirement of the stabilizing function of RecA at 42°C in the presence of rifampin, several factors could be altering the stability of the forks. It is known that high temperatures provoke changes in DNA topology (1, 11). Moreover, rifampin inhibits the initiation of new replication rounds as well as the transcription process, which also alters chromosome supercoiling and nucleoid condensation (6, 7, 39). For these reasons, the structure of reverted forks at 42°C in this context might be especially difficult to maintain. According to this, the environment of the replication fork would be crucial to define the requirement for the stabilizing role of RecA although further studies must be carried out to determine the molecular mechanism of this process.
The results presented here suggest that the absence of RecA impairs the progression of replication when recombination processes are required to restart the stalled forks or that it might even stop DNA synthesis if an additional stabilizing function of the protein was required at the replication forks. This supports the connection between DNA replication and the effect of recombination enzymes on the stalled replication forks at two levels: either by processing stalled forks or maintaining the required DNA structure to be restarted.
ACKNOWLEDGMENTS
We are very grateful to Bénédicte Michel for bacterial strains and experimental support. We also thank Takashi Hishida for the plasmids pRecA and pRecAS25P. We especially thank Alfonso Jiménez-Sánchez for critically reading the manuscript and Encarna Ferrera and María del Carmen Mata Martín for their technical help.
This work was supported by grants PRI07A001 from the Junta de Extremadura and BFU2007-63942 from the Ministerio de Educación y Ciencia. I.S. acknowledges the studentship from the Ministerio de Educación y Cultura FPU AP2002-1157.
Footnotes
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Adamcik J., et al. 2002. Effect of bacteria growth temperature on the distribution of supercoiled DNA and its thermal stability. Electrophoresis 23:3300–3309 [DOI] [PubMed] [Google Scholar]
- 2. Bierne H., Michel B. 1994. When replication forks stop. Mol. Microbiol. 13:17–23 [DOI] [PubMed] [Google Scholar]
- 3. Courcelle J. 2005. Recs preventing wrecks. Mut. Res. 577:217–227 [DOI] [PubMed] [Google Scholar]
- 4. Courcelle J., Ganesan A. K., Hanawalt P. C. 2001. Therefore, what are recombination proteins there for? BioEssays 23:463–470 [DOI] [PubMed] [Google Scholar]
- 5. Courcelle J., Hanawalt P. C. 2003. RecA-dependent recovery of arrested DNA replication forks. Annu. Rev. Genet. 37:611–646 [DOI] [PubMed] [Google Scholar]
- 6. Drlica K., Franco R. J., Steck T. R. 1988. Rifampin and rpoB mutations can alter DNA supercoiling in Escherichia coli. J. Bacteriol. 170:4983–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Drolet M. 2006. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 59:723–730 [DOI] [PubMed] [Google Scholar]
- 8. Eklund H., Uhlin U., Farnegardh M., Logan D. T., Nordlund P. 2001. Structure and function of the radical enzyme ribonucleotide reductase. Prog. Biophys. Mol. Biol. 77:177–268 [DOI] [PubMed] [Google Scholar]
- 9. Fuchs J. A., Karlstrèom H. O., Warner H. R., Reichard P. 1972. Defective gene product in dnaF mutant of Escherichia coli. Nature 238:69–71 [DOI] [PubMed] [Google Scholar]
- 10. Gabbai C. B., Marians K. J. 2010. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair 9:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. González-Soltero R., Botello E., Jiménez-Sánchez A. 2006. Initiation of heat-induced replication requires DnaA and the L-13-mer of oriC. J. Bacteriol. 188:8294–8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grigorian A. V., Lustig R. B., Guzmán E. C., Mahaffy J. M., Zyskind J. W. 2003. Escherichia coli cells with increased levels of DnaA and deficient in recombinational repair have decreased viability. J. Bacteriol. 185:630–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grompone G., Ehrlich D., Michel B. 2004. Cells defective for replication restart undergo replication fork reversal. EMBO Rep. 5:607–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grompone G., Ehrlich S. D., Michel B. 2003. Replication restart in gyrB Escherichia coli mutants. Mol. Microbiol. 48:845–854 [DOI] [PubMed] [Google Scholar]
- 15. Guarino E., Jiménez-Sánchez A., Guzmán E. C. 2007. Defective ribonucleoside diphosphate reductase impairs replication fork progression in Escherichia coli. J. Bacteriol. 189:3496–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guarino E., Salguero I., Jiménez-Sánchez A., Guzmán E. C. 2007. Double-strand break generation under deoxyribonucleotide starvation in Escherichia coli. J. Bacteriol. 189:5782–5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guzmán E. C., Caballero J. L., Jiménez-Sánchez A. 2002. Ribonucleoside diphosphate reductase is a component of the replication hyperstructure in Escherichia coli. Mol. Microbiol. 43:487–495 [DOI] [PubMed] [Google Scholar]
- 18. Horii Z. I., Suzuki K. 1968. Degradation of the DNA of Escherichia coli K12 rec− (JC1569b) after irradiation with ultraviolet light. Photochem. Photobiol. 8:93–105 [DOI] [PubMed] [Google Scholar]
- 19. Horiuchi T., Nishitani H., Kobayashi T. 1995. A new type of E. coli recombinational hotspot which requires for activity both DNA replication termination events and the Chi sequence. Adv. Biophys. 31:133–147 [DOI] [PubMed] [Google Scholar]
- 20. Kogoma T., Cadwell G. W., Barnard K. G., Asai T. 1996. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J. Bacteriol. 178:1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kreuzer K. N. 2005. Interplay between DNA replication and recombination in prokaryotes. Annu. Rev. Microbiol. 59:43–67 [DOI] [PubMed] [Google Scholar]
- 22. Kuzminov A. 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16:373–384 [DOI] [PubMed] [Google Scholar]
- 23. Kuzminov A. 1995. Instability of inhibited replication forks in E. coli. BioEssays 17:733–741 [DOI] [PubMed] [Google Scholar]
- 24. Manwaring J. D., Fuchs J. A. 1979. Relationship between deoxyribonucleoside triphosphate pools and deoxyribonucleic acid synthesis in an nrdA mutant of Escherichia coli. J. Bacteriol. 138:245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Michel B., Boubakri H., Baharoglu Z., LeMasson M., Lestini R. 2007. Recombination proteins and rescue of arrested replication forks. DNA Repair 6:967–980 [DOI] [PubMed] [Google Scholar]
- 26. Michel B., Ehrlich S. D., Uzest M. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Michel B., Grompone G., Flores M.-J., Bidnenko V. 2004. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. U. S. A. 101:12783–12788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miranda A., Kuzminov A. 2003. Chromosomal lesion suppression and removal in Escherichia coli via linear DNA degradation. Genetics 163:1255–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Odsbu I., Morigen, Skarstad K. 2009. A reduction in ribonucleotide reductase activity slows down the chromosome replication fork but does not change its localization. PloS One 4:e7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Riola J., Guarino E., Guzmán E. C., Jiménez-Sánchez A. 2007. Differences in the degree of inhibition of NDP reductase by chemical inactivation and by the thermosensitive mutation nrdA101 in Escherichia coli suggest an effect on chromosome segregation. Cell. Mol. Biol. Lett. 12:70–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sánchez-Romero M. A., Molina F., Jiménez-Sánchez A. 2010. Correlation between ribonucleoside-diphosphate reductase and three replication proteins in Escherichia coli. BMC Mol. Biol. 11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sandler S. J. 2000. Multiple genetic pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics 155:487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sandler S. J., Samra H. S., Clark A. J. 1996. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics 143:5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sassanfar M., Roberts J. W. 1990. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 212:79–96 [DOI] [PubMed] [Google Scholar]
- 35. Seigneur M., Bidnenko V., Ehrlich S. D., Michel B. 1998. RuvAB acts at arrested replication forks. Cell 95:419–430 [DOI] [PubMed] [Google Scholar]
- 36. Shibata T., et al. 2005. Functional overlap between RecA and MgsA (RarA) in the rescue of stalled replication forks in Escherichia coli. Genes Cells 10:181–191 [DOI] [PubMed] [Google Scholar]
- 37. Skarstad K., Boye E. 1993. Degradation of individual chromosomes in recA mutants of Escherichia coli. J. Bacteriol. 175:5505–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Skarstad K., Steen H. B., Boye E. 1985. Escherichia coli DNA distributions measured by flow cytometry and compared with theoretical computer simulations. J. Bacteriol. 163:661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zimmerman S. B. 2006. Shape and compaction of Escherichia coli nucleoids. J. Struct. Biol. 156:255–261 [DOI] [PubMed] [Google Scholar]