Fig. 1.
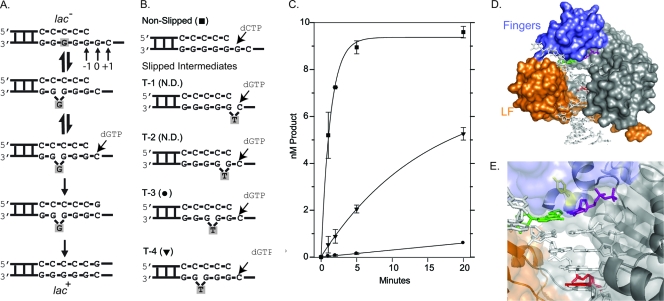
DinB prefers to extend DNA substrates where the “extrahelical” base is located at the −4 position. (A) Using the CC108 lac mutant (lac−) target as an example DNA substrate, the Streisinger template-slippage mechanism is shown. The DNA substrate is in equilibrium between nonslipped and slipped structures (top two structures). The equilibrium can be displaced in favor of the slipped substrate by a dNTP complementary to the +1 base. Further polymerization cycles by DinB will result in the CC108 strain reverting to lac+ as a result of this specific −1 frameshift. The skipped base is shaded throughout the mechanism. Our numbering system is shown. (B) The sequence around the primer terminus of the synthetic oligonucleotide substrates used in our reactions is shown. The primer (20 bp) and the template (33 bp) have the same sequence as the CC108 lac mutant target, except that a dT is positioned within the template strand (indicated) to mimic an extrahelical base. The dNTP used to initiate the reactions are shown. The symbols used to plot the results of the primer extension time course assay in panel C are shown. N.D. indicates that no dNTP incorporation was detected within 20 min. (C) A primer extension time course assay plotting product formation versus time suggests that DinB prefers to extend the T-4 substrate as opposed to the other slipped substrates. (D) A surface representation of our DinB homology model based on a crystal structure of Dpo4 (10). The fingers (blue), little-finger (LF; orange), palm and thumb (gray) domains are shown bound to a DNA substrate. Some solvent-exposed space between the fingers and little-finger domains where the extrahelical base is hypothesized to be located during a template-slippage reaction is visible in the surface representation. (E) A close-up of the active site showing the side chain of Y79 in a stick representation (yellow) as well as the templating base (green), incoming nucleotide (purple), and −4 base (red). The surface representation is semitransparent to reveal the secondary structure of the little-finger domain (orange) as well as that of the active site located in the palm domain. The secondary structure of the fingers domain is not shown for clarity, as it partially encloses the active site.