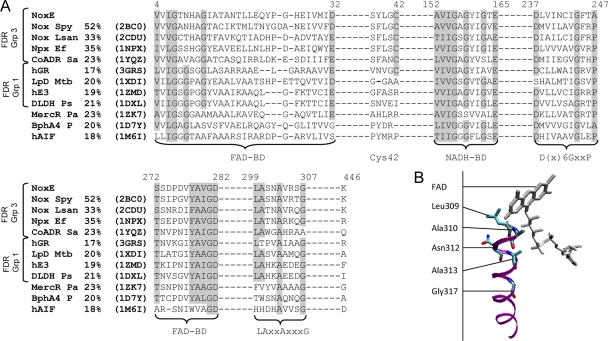
Fig. 2.
(A) Structure-based amino acid alignment of flavoproteins sharing strong structural similarities with L. lactis NoxE. Flavoproteins of the group (Grp) 3 FDR family were as follows: NoxE, NADH oxidase NoxE of L. lactis; Nox Spy, NADH oxidase of S. pyogenes; Nox Lsan, NAD(P)H oxidase of L. sanfranciscensis; Npx Ef, NADH peroxidase of E. faecalis; CoADR Sa, coenzyme A-disulfide reductase of S. aureus. Flavoproteins of the group 1 FDR family were as follows: hGR, Homo sapiens glutathione reductase; LpD Mtb, lipoamide dehydrogenase of Mycobacterium tuberculosis; hE3, human dihydrolipoamide dehydrogenase; DLDH Ps, dihydrolipoamide dehydrogenase of Pisum sativum. The flavoprotein of the group 2 FDR family was MercR Pa, mercuric reductase of P. aeruginosa. Other flavoproteins were as follows: BphA4 P, NADH-dependent ferredoxin reductase component in biphenyl dioxygenase of Pseudomonas sp.; hAIF, human apoptosis induction factor. PDB accession codes are in parentheses. Percentage identities with NoxE are shown in the second column. The positions of amino acid residues of the NoxE sequence are indicated above the sequences. Conserved sequence motifs are shaded. BD, binding domain. (B) Visualization of the alpha-helix formed by residues 309 to 317 and FAD in the structural model of S. pyogenes NoxE (PDB code 2BC0).