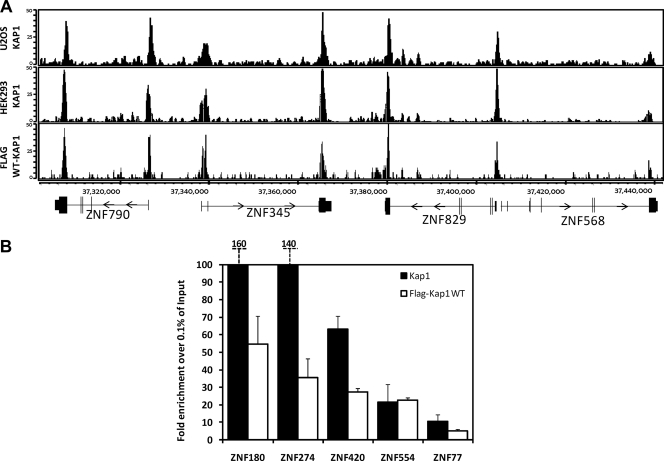
Fig. 2.
Comparison of FLAG-tagged and endogenous KAP1. (A) ChIP-seq binding patterns. Snapshots of ChIP-seq results for endogenous KAP1 in U2OS cells, endogenous KAP1 in HEK293 cells, and FLAG-tagged WT KAP1 in HEK293 cells for a region on chromosome 19. The number of sequenced tags is shown on the y axis, and the coordinates of several ZNF genes are shown on the x axis. (B) ChIP-PCR confirmation of KAP1 binding sites. ChIPs were performed with HEK293 cells stably expressing FLAG-tagged WT KAP1. Antibodies that recognize KAP1 or the FLAG tag were used. Five binding sites identified by ChIP-seq were confirmed by qPCR using independent ChIP samples. The fold enrichment of KAP1 and FLAG-tagged WT KAP1 (compared to 0.1% of the starting chromatin) is shown; results are from two independent replicates; the error bars indicate standard errors.