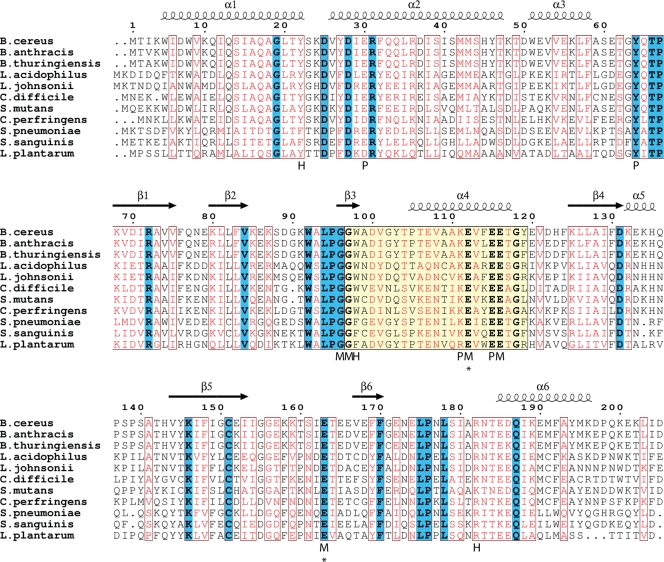
Fig. 3.
Family of CDP-Chase-like enzymes. A BLAST search reveals many putative Nudix hydrolases with a high degree of sequence similarity to CDP-Chase. Identical residues are in boldface and highlighted in blue, and residues with high similarity are shown in red. The Nudix signature sequence is highlighted in yellow. The secondary structural assignment above the sequences corresponds to that of monomer A of CDP-Chase. Below the sequences, residues are labeled according to their predicted interactions with CDP-choline: H marks residues that are within hydrogen bonding distance, M marks residues that either coordinate the catalytic Mg2+ or interact with these residues through solvent-mediated interactions, and P marks other residues that line the binding pocket of CDP-choline. Asterisks indicate residues that were mutated to study activity. B. thuringiensis, Bacillus thuringiensis; L. acidophilus, Lactobacillus acidophilus; L. johnsonii, Lactobacillus johnsonii; C. difficile, Clostridium difficile; S. mutans, Streptococcus mutans; C. perfringens, Clostridium perfringens; S. sanguinis, Streptococcus sanguinis; L. plantarum, Lactobacillus plantarum.