Fig. 4.
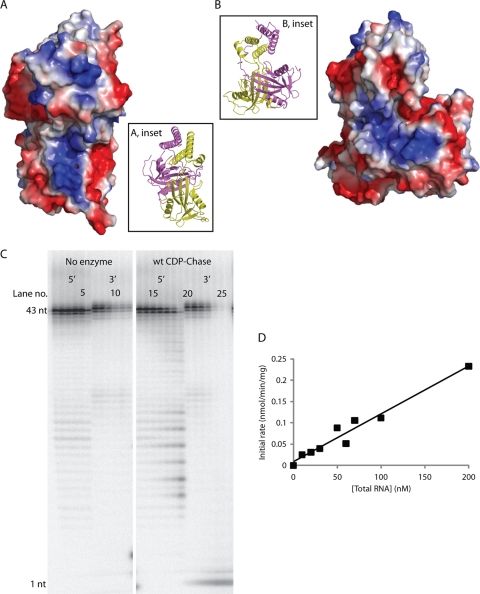
CDP-Chase has an RNA exonuclease activity. CDP-Chase contains two positively charged grooves, one on each monomer, which may facilitate RNA binding. (A) Electrostatic surface of CDP-Chase centered on the positively charged electrostatic groove of monomer A. (Inset) Ribbon diagram of protein in the same orientation as the electrostatic surface. The coloring of the protein backbone is the same as in Fig. 2A. (B) Electrostatic surface of CDP-Chase centered on the positively charged electrostatic groove of monomer B. (Inset) Ribbon diagram of protein in the same orientation as the electrostatic surface. (C) To identify any RNase activity by B. cereus CDP-Chase, an RNase assay was performed on RNA labeled with 32P-monophosphate on either the 5′ end or the 3′ end in the absence (lanes 1 to 6 for 5′-end-labeled RNA and lanes 7 to 12 for 3′-end-labeled RNA) or presence (lanes 13 to 19 for 5′-end-labeled RNA and lanes 20 to 26 for 3′-end-labeled RNA) of enzyme. In the assay with no enzyme, the two sets of six lanes represent time points of 0, 5, 10, 20, 30, and 60 min, from left to right. In the assay with enzyme (wt CDP-Chase), the two sets of seven lanes represent time points of 1, 2, 5, 10, 20, 30, and 60 min, from left to right. wt, wild type; nt, nucleotide. (D) Competition assay of RNA degradation by CDP-Chase. The data were fit to a linear model.