Fig. 2.
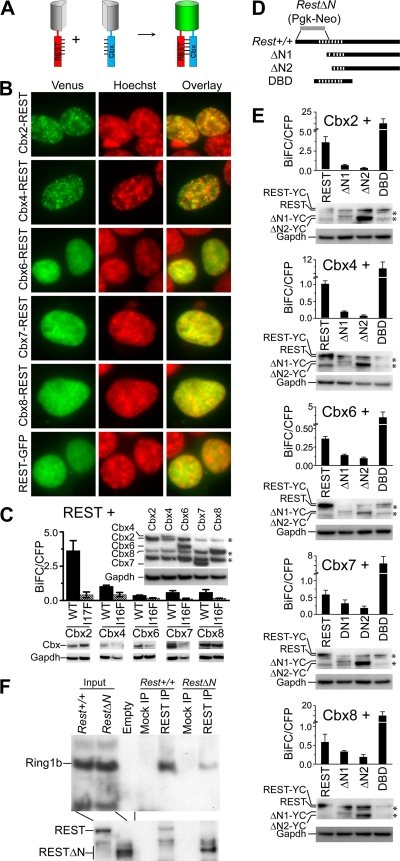
REST interactions with Cbx proteins in living cells and with Ring1b in native cell extracts. (A) Principle for BiFC analysis of REST interactions with Cbx proteins. REST and Cbx proteins fused to fluorescent protein fragments were coexpressed. The interaction of REST with Cbx proteins was detected based on the facilitated association of the fluorescent protein fragments. (B) The subnuclear distributions of complexes formed by the Cbx family proteins and REST indicated at the left of the images were visualized by using BiFC analysis (green) in live HEK293T cells stained by Hoechst dye (red). Different exposure times were used to compensate for differences in fluorescence intensities of BiFC complexes formed by different Cbx proteins with REST. The images are representative of the majority of cells in each population. (C) Efficiencies of BiFC complex formation by REST with wild-type (WT) and mutant (I17F or I16F) Cbx proteins measured by flow cytometry. The bars show the mean fluorescence intensities of 20,000 cells in each of two independent experiments. The levels of expression of different Cbx fusion proteins were compared by analyzing equal amounts of the cell extracts by immunoblotting using anti-GFP antibodies (inset above bars) (*, cross-reactive proteins). The levels of wild-type and mutant Cbx fusion protein expression were similarly compared (below the graph). The levels of Gapdh were measured to determine the amount loaded into each lane. (D) The diagrams indicate the truncated REST variants used to map interactions with Cbx proteins. The stripes indicate the zinc finger DNA-binding domain. The region replaced by Pgk-Neo in RestΔN knockout cells is indicated by a gray bar (9). (E) Efficiencies of BiFC complex formation by the Cbx proteins indicated above the bars with the truncated REST proteins indicated below the bars measured by flow cytometry. The bars show the mean fluorescence intensities of 20,000 cells in each of two independent experiments. The levels of expression of full-length and truncated REST fusions were measured by immunoblotting using anti-REST antibody (below each graph) (*, cross-reactive proteins). Gapdh was measured to determine the amount loaded into each lane. (F) Analysis of endogenous REST and Ring1b interactions in ES cell extracts. Extracts from Rest+/+ and RestΔN ES cells were precipitated by using anti-REST antibodies or beads alone (mock). The precipitates were analyzed by immunoblotting using antibodies directed against the proteins indicated at the left of the image. The input lanes contained 5% of the extract.