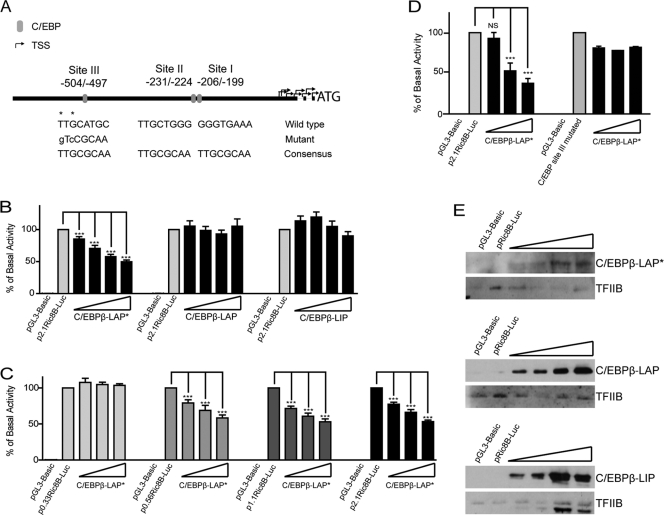
Fig. 4.
C/EBPβ-LAP* negatively regulates the Ric-8B promoter activity in osteoblastic cells. (A) Schematic diagram of putative C/EBP elements within the first 600 bp of the human, mouse, and rat Ric-8B promoters. Negative values indicate the position of these sites relative to ATG. To evaluate the contribution of C/EBP site III, nucleotides marked with asterisks were replaced by the nucleotides showed in lowercase letters by using site-directed mutagenesis. (B) Subconfluent MC3T3 cells were cotransfected with the construct p2.1Ric-8B-Luc (100 ng) and increasing amounts (100 to 800 ng) of the plasmids encoding each C/EBPβ isoform: C/EBPβ-LAP*, C/EBPβ-LAP, and C/EBPβ-LIP. Twenty-four hours later, luciferase activity was determined. (C) MC3T3 cells were cotransfected with the different constructs containing the Ric-8B promoter sequences (2.1, 1.1, 0.56, and 0.33 kb) and increasing amounts of the plasmid encoding C/EBPβ-LAP* (200 to 800 ng). Twenty-four hours after transfection, luciferase activity was determined. (D) MC3T3 cells were cotransfected with the p2.1Ric-8B-Luc plasmid, including a mutated putative C/EBP site III (p2.1Ric-8B-MutC/EBPIII-Luc; 200 ng) and increasing amounts (100 to 800 ng) of the plasmid encoding for C/EBPβ-LAP*. At 24 h after transfection, luciferase activity was determined and compared with that of the wild-type p2.1Ric-8B-Luc promoter construct. (E) Ten micrograms of nuclear extracts obtained from MC3T3 cells transfected under the conditions described for panel B was analyzed by Western blotting to control for overexpression of C/EBPβ. Reblotting against TFIIB was used to control for equal loading. The graphs show the percentage of basal activity, calculated as the ratio between firefly and Renilla luciferase activities. The bars represent the means ± standard errors of the means of three independent experiments, each performed in triplicate. Statistically significant differences were determined by the analysis of variance test. NS, not significant; ***, P < 0.001.