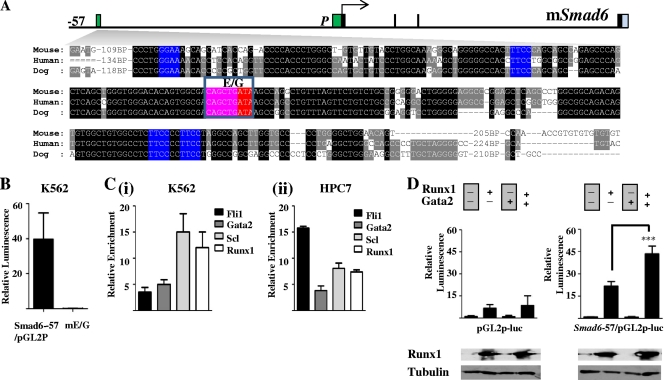
Fig. 3.
Smad6-57 region has conserved Ets, Gata, and E-box motifs, is active in hematopoietic cells, and can be transactivated by Runx1 in the absence of a Runx1 binding motif. (A) Nucleotide sequence alignment of the kb −57 mSmad6 (homologous to the kb −71 hSMAD6) region. Conserved Ets (E), Gata (G), and E-box (EB) motifs are colored in blue, red, and pink, respectively. (B) A 500-bp fragment from the kb −57 mSmad6 region (−57) is active in K562 cells. Stable transfection assays in K562 cells with wild-type and E-box/Gata-mutated (mE/G) fragments subcloned into the pGL2p vector. The luciferase activities are given as the fold increase above the activity of the pGL2p vector. Each bar is the means (± standard deviations) of the relative luciferase activity from at least two experiments performed in triplicate. (C) ChIP assays in human and mouse cell lines. (i) The hSMAD6 −71 enhancer is bound by SCL, FLI1, and GATA2 in K562 erythroleukemic cells. (ii) The mSmad6-57 enhancer also is bound by Scl, Fli1, and Gata2 in HPC-7 stem/progenitor cells. (D) The mSmad6-57 enhancer (Smad6-57/pGL2p-luc) can be transactivated by Runx1, but this effect is greatly enhanced by Gata2. The pGL2p-luc vector was used as a control. ***, P < 0.001.