Fig. 4.
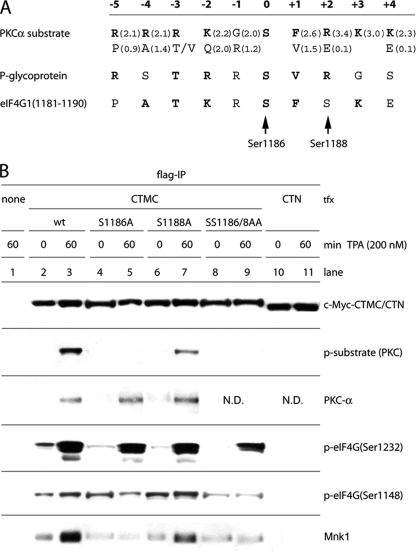
eIF4G(Ser1186) is phosphorylated by PKCα. (A) Optimal substrate consensus for PKCα, sequences of a confirmed substrate (P-glycoprotein of MDR1) (37) and of the proposed phosphorylation site in eIF4G (aa 1179 to 1190). Values in parentheses indicate the relative preference for the amino acids shown (bold letters represent amino acids with values of >1.5). Bold letters in the eIF4G sequence emphasize positions that are equal to the consensus or to the P-glycoprotein phosphorylation site. (B) Identification of the PKCα phosphorylation site in eIF4G. HEK293 cells were transfected for 16 h with CTMC variants or CTN fragments, serum starved for 24 h, and treated for 60 min with DMSO (0) or 200 nM TPA. Cell lysates were subjected to Flag-IP followed by immunoblotting. This assay was repeated three times; a representative experiment is shown. ND, not done.