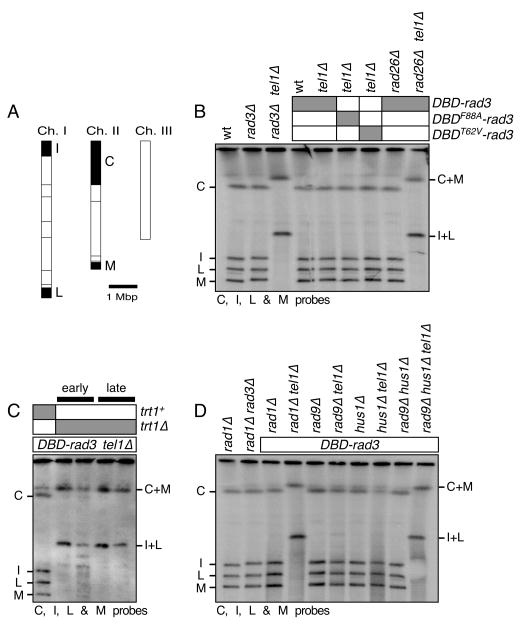
Figure 3.
PFGE analysis of NotI digested chromosomal DNA for the indicated DBD-rad3 related strains. (A) NotI restriction enzyme map of fission yeast chromosomes (horizontal lines). The telomeric probes C, I, L and M, used in Southern blot hybridization are filled black and marked. Chromosome circularization is indicated by the appearance of C+M and I+L fusion bands. Chromosome III was not monitored since it lacks NotI site. (B-D) Chromosomal DNA was prepared in agarose plugs, digested with NotI, and then used in PFGE for the indicated strains, which were extensively restreaked on agar plates to ensure terminal telomere phenotype. DNA was then transferred to a Nylon membrane and hybridized to C, I, L and M specific probes. For (C), “early” samples are prepared from confirmed trt1Δ strains immediately after transformation with the trt1Δ construct, while “late” samples are prepared from strains restreaked four times on agar plates after transformation with the trt1Δ construct.