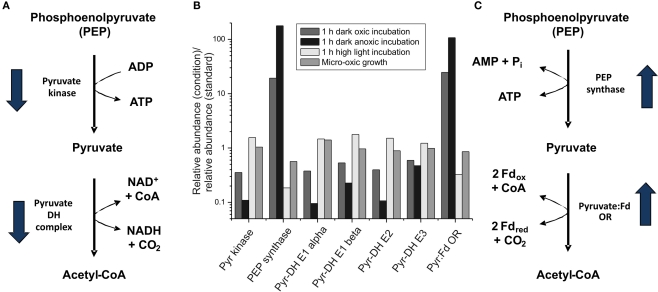
Figure 6.
Changes in the transcript levels for genes coding for proteins involved in pyruvate metabolism. Relative transcript levels for genes encoding proteins/enzymes involved in pyruvate metabolism [pyruvate (Pyr) kinase (pyk), phosphoenolpyruvate (PEP) synthase (ppsA), pyruvate dehydrogenase (Pyr-DH) complex (E1 alpha, E1 beta, E2 and E3 proteins; encoded by pdhA, pdhB, SYNPCC7002_A0110, ipdA), and pyruvate:ferredoxin oxidoreductase (Pyr:Fd OR; nifJ)] are shown. The proposed reaction pathways from PEP to acetyl-CoA with the respective enzymes are shown in (A,C); and the change in the mRNA level for the respective genes upon dark incubation is indicated by arrows. (B) The ratio of the transcript levels for cells after a 1-h dark oxic treatment (dark gray), a 1-h dark anoxic treatment (fermentative conditions; black), a 1-h high-light treatment (light gray), and photolithoautotrophic growth under micro-oxic conditions (medium gray) are compared to transcript levels in cells under standard conditions.