Abstract
Circoviruses consist of highly prevalent and genetically diverse porcine and avian pathogens. The genomes of cycloviruses, a proposed new genus in the family Circoviridae, were recently identified in human and chimpanzee faeces. Here, six cyclovirus and four circovirus genomes from the tissues of chickens, goats, cows, and a bat were amplified and sequenced using rolling-circle amplification and inverse PCR. A goat cyclovirus was nearly identical to a cyclovirus from a cow. USA beef contained circoviruses with >99 % similarity to porcine circovirus 2b. Circoviruses in chicken were related to those of pigeons. The close genetic similarity of a subset of cycloviruses and circoviruses replicating in distinct animal species may reflect recent cross-species transmissions. Further studies will be required to determine the impact of these highly prevalent infections on the health of farm animals.
Members of the family Circoviridae, are non-enveloped, spherical viruses with a single-stranded circular DNA genome of approximately 2 kb, the smallest known autonomously replicating viral genomes (Todd, 2005). Circoviruses cause a variety of clinical symptoms in birds and pigs including lethargy, lymphoid depletion and immunosuppression (Hattermann et al., 2003; Johne et al., 2006a; Mankertz et al., 2000; Niagro et al., 1998; Shivaprasad et al., 2004; Soike et al., 2004; Stewart et al., 2006; Todd, 2005; Todd et al., 2001).
Both circoviruses and the recently described cycloviruses have an ambisense genome organization containing two major inversely arranged ORFs encoding the putative replication-associated protein (Rep) and capsid protein (Cap). A potential stem–loop structure with a conserved nonanucleotide motif located between the 5′-ends of these two ORFs is required to initiate the replication of the viral genome (Hattermann et al., 2003; Johne et al., 2006a; Mankertz et al., 2000; Niagro et al., 1998; Stewart et al., 2006; Todd, 2005; Todd et al., 2001, 2007). Cycloviruses are distinguishable from circoviruses by missing one intergenic region, containing a different conserved nonamer sequence atop their stem–loop structure and by phylogenetically clustering separately from the circoviruses.
To date no circovirus infection has been reported in chickens, and porcine circoviruses 1 and 2 (PCV1 and PCV2) are the only two circoviruses reported to infect mammals. PCV2 was reported to be present in cattle with respiratory disease and from aborted bovine fetuses (Nayar et al., 1999), but this finding has not been subsequently confirmed (Rodríguez-Arrioja et al., 2003). A more recent study of calves in Germany with a fatal haemorrhagic disease syndrome also detected PCV2 (Kappe et al., 2010).
Using degenerate PCR primers based on highly conserved amino acid motifs in the Rep proteins, both circovirus and cyclovirus related sequences were recently detected in muscle tissues of animals including chickens, cows, sheep, goats and camels from Pakistan and Nigeria (Li et al., 2010a), but complete genomes were not obtained from these tissues. In this study, rolling-circle amplification, using the illustra TempliPhi 100 Amplification kit (GE Health Care) according to a modified protocol optimized for the amplification of viral circular DNA genomes (Johne et al., 2006b; Stevens et al., 2010), and inverse PCR were performed to amplify and sequence some of these viruses. We acquired the complete genome sequences of 11 circoviruses, cycloviruses and a circovirus-like virus from farm animals from Pakistan, Nigeria and the USA and from a bat from Texas. We also extended PCR prevalence measurements to include chicken, pork and beef from stores in California, USA (GenBank accession nos HQ738634–HQ738644 and HQ839714–HQ839728). The sampling place and time of the Pakistani and Nigerian farm animal tissue samples were reported previously (Li et al., 2010a). The USA meat samples (chicken, beef and pork) were collected from California, USA in September 2008, and from October 2009 to July 2010.
We previously reported that none of the 13 Pakistani, but 30 of the 40 Nigerian chicken muscle tissue samples were PCR positive for the Rep gene (Li et al., 2010a). Twenty-two of the 30 Nigerian sequences were closely related and belonged to the genus Cyclovirus, while eight sequences clustered with columbid circovirus (CoCV) from pigeons. All 21 San Francisco supermarket-bought chicken samples were negative for circovirus-like Rep sequence.
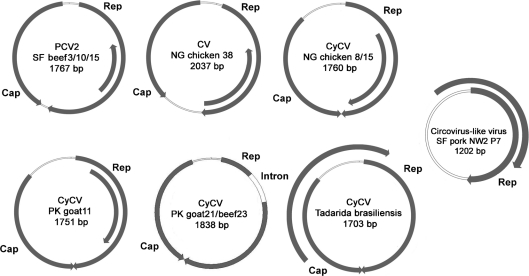
The full-length genomes of two closely related chicken cycloviruses (CyCV-NG chicken 8 and CyCV-NG chicken 15) and that of the chicken circovirus (CV-NG chicken 38) were obtained. Sequence analysis revealed that the cycloviruses were 1760 nt long and circular, and shared 99 % nucleotide identity differing at only 10 nt, resulting in one amino acid change in the Rep and the Cap proteins. The chicken circovirus genome was 2037 nt and was 92 % identical to CoCV.
The genome of the chicken cycloviruses and circovirus had features characteristic of their genera, including the absence and presence, respectively, of an intergenic region between the 3′ of both major ORFs (Li et al., 2010a; Todd, 2005) (Fig. 1). CV-NG chicken 38 had the typical stem–loop structure and nonamer sequence of circoviruses (5′-TAGTATTAC-3′), while the nonamer sequence of chicken cycloviruses (5′-TAATACTAA-3′) slightly differed from those of cycloviruses in human and chimpanzee faeces (5′-TAATACTAT-3′) (Li et al., 2010a). No suitably located ATG was identified for the ORF encoding the Cap protein of CV-NG chicken 38. Considering the common use of alternative start codons in avian circoviruses (Niagro et al., 1998; Phenix et al., 2001; Stewart et al., 2006; Todd et al., 2001, 2007), ATA was considered as the most likely start codon producing a Cap ORF of comparable size with that of CoCV. CoCV uses the alternative start codon ATA for both the Rep and Cap ORFs.
Fig. 1.
Genomic organization of the cycloviruses, circoviruses and circovirus-like virus discovered in animal tissues. The two major ORFs, encoding the putative Rep protein and the putative Cap protein, and other ORFs with a coding capacity over 100 aa are shown.
The putative Rep proteins of the chicken cycloviruses were 278 aa and from 47 to 74 % similar to the Rep of previously reported cycloviruses (Li et al., 2010a, b) (Supplementary Table S1, available in JGV Online). The deduced Cap proteins were 222 aa long, typical of cycloviruses (average 220 aa), exhibiting 14–48 % similarity with those of reported cycloviruses (Supplementary Table S2, available in JGV Online). The chicken circovirus had a Rep protein of 317 aa, as does CoCV, with which it shared 93 % amino acid identity (Supplementary Table S1). The Cap protein of the chicken circovirus was 273 aa, also the same length as CoCV, with an amino acid similarity of 98 %.
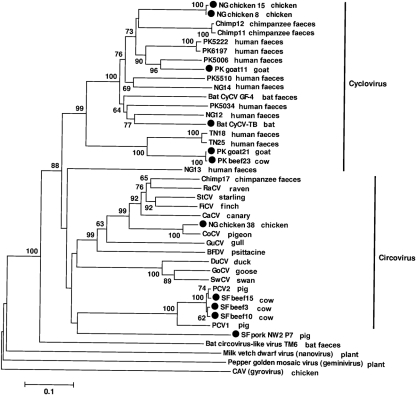
A phylogenetic analysis of Rep confirmed that CyCV-NG chicken 8/15 clustered with cycloviruses, while CV-NG chicken 38 was closely related to CoCV (Mankertz et al., 2000; Todd et al., 2008) (Fig. 2).
Fig. 2.
Phylogenetic analysis of the cycloviruses, circoviruses and circovirus-like virus, and representative cyclovirus and circovirus species based on the complete amino acid sequence of the Rep protein using the neighbour-joining method with p-distance and 1000 bootstrap replicates. The bar represents 10 % estimated phylogenetic divergence. The GenBank accession numbers of the Rep sequences of viruses used in the phylogenetic analyses are as follows: beak and feather disease virus (BFDV, AF071878), canary circovirus (CaCV, AJ301633), CoCV (AF252610), duck circovirus (DuCV, DQ100076), goose circovirus (GoCV, AJ304456), gull circovirus (GuCV, DQ845074), finch circovirus (FiCV, DQ845075), raven circovirus (RaCV, DQ146997), starling circovirus (StCV, DQ172906), cygnus olor circovirus (SwCV, EU056310), PCV1 (AY660574), PCV2 (AY424401), chicken anemia virus (CAV, M55918), milk vetch dwarf virus (AB009047), pepper golden mosaic virus (U57457), cycloviruses (GQ404844–GQ404850, GQ404854–GQ404858, HM228874 and HM228875).
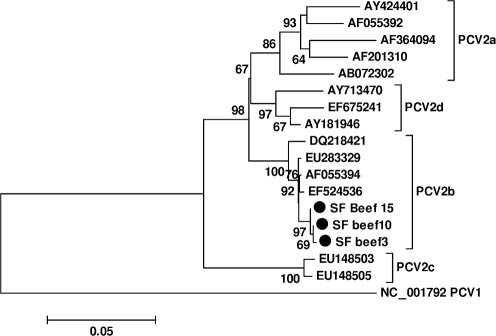
Circovirus-like Rep sequences were detected in 9 of 19 beef samples from supermarkets in San Francisco, USA. Five were PCV2 sequences, and four were similar to the previously reported circovirus-like sequence NW2 with higher than 96 % nucleotide identity over a Rep fragment of approximately 400 nt amplified by degenerate PCR (Li et al., 2010a). Including 51 beef specimens from Pakistan and Nigeria tested in a previous study (Li et al., 2010a), there were 17 of 70 positive beef specimens, with seven cyclovirus sequences, five PCV2 sequences and five circovirus-like sequences. We obtained the full-length genomes of PCV2 from three USA beef specimens (PCV2 SF beef3, 10 and 15). The three PCV2 from beef were 1767 nt in length, differing at 5, 14 and 15 nt, respectively, with one another and sharing 99 % nucleotide identity with PCV2 strains from pigs. Phylogenetically all clustered with the PCV2b genotypes (Grau-Roma et al., 2008; Guo et al., 2010) (Fig. 3), consistent with the recent finding of PCV2b in calves with a haemorrhagic disease syndrome in Germany (Kappe et al., 2010). One cyclovirus from beef was also sequenced (CyCV-PK beef23).
Fig. 3.
PCV2 genotypes. Phylogenetic analysis was based on the nucleotide sequences of the full-length ORF2 of representative PCV2 strains.
In the previous study, 8 of 73 goats and sheep specimens from Pakistan and Nigeria were PCR positive for Rep, all from the genus Cyclovirus (Li et al., 2010a). Two cyclovirus genomes were obtained from Pakistani goats (CyCV-PK goat11 and CyCV-PK goat21). The genomes of CyCV-PK goat21 and CyCV-PK beef23 were both 1838 nt, shared 99 % nucleotide identity, differing only at 2 nt resulting in 1 aa difference in Rep (Fig. 1). The Rep gene of CyCV-PK goat21 and CyCV-PK beef23 were both interrupted by a 169 bp intron with typical splice donor (GT) and splice acceptor site (AG) as were the related human faeces-derived CyCV-TN18 and CyCV-TN25 (Li et al., 2010a). Both Rep proteins were 280 aa, sharing 48–75 % similarity with known cycloviruses (Li et al., 2010a) (Supplementary Table S1). The deduced Cap proteins were 212 aa, showing 14–57 % similarity with those of known cycloviruses and having the highest similarity of 57 % with CyCV-TN18 and CyCV-TN25 (Supplementary Table S2). The stem–loop structure contained a slightly modified nonamer sequence (5′-TAATACTAG-3′) compared with cycloviruses identified in human and chimpanzee faeces (5′-TAATACTAT-3′). Phylogenetically, CyCV-PK goat21 and CyCV-PK beef23 clustered with human faeces derived CyCV-TN18 and CyCV-TN25 (Fig. 2).
The CyCV-PK goat11 genome was 1751 nt long encoding a 278 aa Rep and a 231 aa Cap protein (Fig. 1). Its nonanucleotide motif (5′-TAATACTAT-3′) was the same as the chimpanzee and human stool CyCV. The Rep of CyCV-PK goat11 showed 83 % similarity with a human faeces derived CyCV-PK5006, while Cap showed <42 % amino acid similarity with other cycloviruses (Supplementary Tables S1 and S2). Phylogenetically, CyCV-PK goat11 distantly clustered with other CyCV from human Pakistani faeces (Fig. 2).
In total, 18 of 22 pork products (muscle, ground pork and ham) from San Francisco supermarkets were PCR positive for circovirus-like Rep sequence (Li et al., 2010a). Twelve samples were positive for PCV2, and one sample was positive for PCV1. Six samples were closely related to one another and to the previously reported circovirus-like virus pork NW2 sequence (Li et al., 2010a). In one pork sample, both PCV2 and NW2 were detected. We obtained the full-length genomes of circovirus-like virus SF pork NW2 P7 and the partial genomes of closely related strains pork NW2 P8 (1050 bp) and NW2 P9 (899 bp). All shared more than 97 % nucleotide identity with each other.
Circovirus-like virus SF pork NW2 P7 genome was only 1202 nt and circular, much shorter than known circoviruses and cycloviruses. Its genome organization resembled those of the single-stranded circular DNA anelloviruses with two overlapping ORFs in the same direction (Fig. 1), but with a shorter genome (Biagini, 2009). No suitably located ATG was identified for ORF1 encoding the putative Rep protein, but GTC was considered a possible alternative. The initiation codon for the putative ORF2 was ATG. The putative Rep protein of circovirus-like virus SF pork NW2 P7 was 221 aa, with 34–46 % similarity to the Rep protein of known cycloviruses, and 39–46 % similarity to circoviruses (Supplementary Table S1). ORF2 encoded a putative protein of 177 aa, which had low amino acid similarity (41 %) with a hypothetical protein m169 of muromegalovirus, a member of the family Herpesviridae. A stem–loop structure was found 51 nt upstream of ORF2 with no homology to those of circoviruses or cycloviruses. Phylogenetic analyses of its circovirus-like Rep proteins showed that it fell outside the circovirus group, but grouped together with the combined circoviruses and cyclovirus clades (Fig. 2).
The pectoral muscle, digestive tract and faecal specimen from an individual male Brazilian free-tailed bat (Tadarida brasiliensis) in Texas, USA, in 2009 were all PCR positive for circovirus-like Rep sequence. The three sequences were identical with one another and belonged to the cyclovirus clade, indicating that this virus infected this bat rather than simply being consumed and excreted. The full-length genome of this cyclovirus was sequenced from muscle tissue, and tentatively named cyclovirus Tadarida brasiliensis (CyCV-TB).
The CyCV-TB genome was 1703 nt, with a typical cyclovirus genome organization (Fig. 1). The putative Rep protein of CyCV-TB was 278 aa, with 44–71 % similarity to the Rep protein of known cycloviruses. CyCV-TB showed the highest amino acid similarity of 71 % to CyCV-NG12 from a Nigerian human faeces, and 68 % amino acid similarity with the CyCV-GF4 genome previously reported in bat guano from a Californian roost (Li et al., 2010b) (Supplementary Table S1). The deduced Cap protein was 225 aa, showing 12–48 % similarity with those of cycloviruses found in human and chimpanzee, and 28 % amino acid similarity with the bat guano derived CyCV-GF4 (Supplementary Table S2). The highly conserved stem–loop structure with the nonamer sequence (5′-TAATACTAT-3′), identical to that in cycloviruses from human and chimpanzee faeces, was present in the 5′-intergenic region.
The International Committee for the Taxonomy of Viruses suggested criteria for circovirus species demarcation as genome nucleotide identities of less than 75 % and Cap protein amino acid identities of less than 70 % (Todd, 2005). This study reports on circular ssDNA viruses in the tissue of farm animals and a wild bat. Based on the distance criteria, CV-NG chicken 38 therefore appears to be a subtype of CoCV, and SF beef3, 10 and 15 are strains of PCV2b. Four novel species of cycloviruses were characterized, including CyCV-NG chicken 8/15, CyCV-PK goat11, CyCV-PK goat21/beef23 and CyCV-TB. Circovirus-like SF pork NW2 P7 genome is unusually small and only loosely related to circoviruses or cycloviruses and because of its unusual genome size and organization, its classification remains uncertain. The detection of apparently truncated circular DNA genome is reminiscent of that reported for a distantly related group of circular DNA viruses (ChiSCV) recently detected in chimpanzee stool (Blinkova et al., 2010) and could reflect the presence of defective genome requiring trans-complementation by a helper virus.
Infection of different animal species by very closely related viruses includes PCV2 in pork and beef, CoCV in pigeon and chicken, CyCV-PK goat21/beef23 in goat and cow, and circovirus-like virus SF pork NW2 in pork and beef. Given that circoviruses have been estimated to have a rate of nucleotide substitution approaching those of RNA viruses (Firth et al., 2009), the presence of closely related viruses in different hosts may reflect recent cross-species transmissions.
Beside the well documented pathogenicity of PCV2 in pigs and of other circoviruses in birds, the biological significance of widespread cyclovirus, circovirus and circovirus-like virus infections in other domesticated and wild animals remains unknown. Further epidemiological and pathogenesis studies will be required to assess the significance of these common infections on these important sources of human dietary proteins.
Supplementary Material
Acknowledgments
The work was supported by the Blood Systems Research Institute and NIH R01 HL083254 to Dr Eric Delwart.
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the sequences reported in this study are HQ738634–HQ738644 and HQ839714–HQ839728.
Supplementary tables are available with the online version of this paper.
References
- Biagini, P. (2009). Classification of TTV and related viruses (anelloviruses). Curr Top Microbiol Immunol 331, 21–33. [DOI] [PubMed] [Google Scholar]
- Blinkova, O., Victoria, J., Li, Y., Keele, B., Sanz, C., Ndjango, J. B., Peeters, M., Travis, D., Lonsdorf, E. & other authors (2010). Novel circular DNA viruses in stool samples of wild-living chimpanzees. J Gen Virol 91, 74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth, C., Charleston, M. A., Duffy, S., Shapiro, B. & Holmes, E. C. (2009). Insights into the evolutionary history of an emerging livestock pathogen: porcine circovirus 2. J Virol 83, 12813–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau-Roma, L., Crisci, E., Sibila, M., López-Soria, S., Nofrarias, M., Cortey, M., Fraile, L., Olvera, A. & Segalés, J. (2008). A proposal on porcine circovirus type 2 (PCV2) genotype definition and their relation with postweaning multisystemic wasting syndrome (PMWS) occurrence. Vet Microbiol 128, 23–35. [DOI] [PubMed] [Google Scholar]
- Guo, L. J., Lu, Y. H., Wei, Y. W., Huang, L. P. & Liu, C. M. (2010). Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J 7, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattermann, K., Schmitt, C., Soike, D. & Mankertz, A. (2003). Cloning and sequencing of Duck circovirus (DuCV). Arch Virol 148, 2471–2480. [DOI] [PubMed] [Google Scholar]
- Johne, R., Fernández-de-Luco, D., Höfle, U. & Müller, H. (2006a). Genome of a novel circovirus of starlings, amplified by multiply primed rolling-circle amplification. J Gen Virol 87, 1189–1195. [DOI] [PubMed] [Google Scholar]
- Johne, R., Wittig, W., Fernández-de-Luco, D., Höfle, U. & Müller, H. (2006b). Characterization of two novel polyomaviruses of birds by using multiply primed rolling-circle amplification of their genomes. J Virol 80, 3523–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappe, E. C., Halami, M. Y., Schade, B., Alex, M., Hoffmann, D., Gangl, A., Meyer, K., Dekant, W., Schwarz, B. A. & other authors (2010). Bone marrow depletion with haemorrhagic diathesis in calves in Germany: characterization of the disease and preliminary investigations on its aetiology. Berl Munch Tierarztl Wochenschr 123, 31–41. [PubMed] [Google Scholar]
- Li, L., Kapoor, A., Slikas, B., Bamidele, O. S., Wang, C., Shaukat, S., Masroor, M. A., Wilson, M. L., Ndjango, J. B. & other authors (2010a). Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol 84, 1674–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Victoria, J. G., Wang, C., Jones, M., Fellers, G. M., Kunz, T. H. & Delwart, E. (2010b). Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol 84, 6955–6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankertz, A., Hattermann, K., Ehlers, B. & Soike, D. (2000). Cloning and sequencing of columbid circovirus (CoCV), a new circovirus from pigeons. Arch Virol 145, 2469–2479. [DOI] [PubMed] [Google Scholar]
- Nayar, G. P., Hamel, A. L., Lin, L., Sachvie, C., Grudeski, E. & Spearman, G. (1999). Evidence for circovirus in cattle with respiratory disease and from aborted bovine fetuses. Can Vet J 40, 277–278. [PMC free article] [PubMed] [Google Scholar]
- Niagro, F. D., Forsthoefel, A. N., Lawther, R. P., Kamalanathan, L., Ritchie, B. W., Latimer, K. S. & Lukert, P. D. (1998). Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch Virol 143, 1723–1744. [DOI] [PubMed] [Google Scholar]
- Phenix, K. V., Weston, J. H., Ypelaar, I., Lavazza, A., Smyth, J. A., Todd, D., Wilcox, G. E. & Raidal, S. R. (2001). Nucleotide sequence analysis of a novel circovirus of canaries and its relationship to other members of the genus Circovirus of the family Circoviridae. J Gen Virol 82, 2805–2809. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Arrioja, G. M., Segalés, J., Domingo, M. & Plana-Duran, J. (2003). Lack of PCV-2 infection in non-porcine species in Spain. Vet Rec 153, 371–372. [PubMed] [Google Scholar]
- Shivaprasad, H. L., Hill, D., Todd, D. & Smyth, J. A. (2004). Circovirus infection in a Gouldian finch (Chloebia gouldiae). Avian Pathol 33, 525–529. [DOI] [PubMed] [Google Scholar]
- Soike, D., Albrecht, K., Hattermann, K., Schmitt, C. & Mankertz, A. (2004). Novel circovirus in mulard ducks with developmental and feathering disorders. Vet Rec 154, 792–793. [DOI] [PubMed] [Google Scholar]
- Stevens, H., Rector, A. & Van Ranst, M. (2010). Multiply Primed Rolling-Circle Amplification Method for the Amplification of Circular DNA Viruses, pdb prot5415. Cold Spring Harbor, NY. : Cold Spring Harbor Protocols. [DOI] [PubMed]
- Stewart, M. E., Perry, R. & Raidal, S. R. (2006). Identification of a novel circovirus in Australian ravens (Corvus coronoides) with feather disease. Avian Pathol 35, 86–92. [DOI] [PubMed] [Google Scholar]
- Todd, D. (2005). Circoviridae. In Virus Taxonomy: the Eighth Report of the International Committee on Taxonomy of Viruses, pp. 326–334. Edited by Fauquet, C. M., Mayo, M. A., Maniloff, J., Desselberger, U. & Ball, L. A.. New York. : Academic Press.
- Todd, D., Weston, J. H., Soike, D. & Smyth, J. A. (2001). Genome sequence determinations and analyses of novel circoviruses from goose and pigeon. Virology 286, 354–362. [DOI] [PubMed] [Google Scholar]
- Todd, D., Scott, A. N., Fringuelli, E., Shivraprasad, H. L., Gavier-Widen, D. & Smyth, J. A. (2007). Molecular characterization of novel circoviruses from finch and gull. Avian Pathol 36, 75–81. [DOI] [PubMed] [Google Scholar]
- Todd, D., Fringuelli, E., Scott, A. N., Borghmans, B. J., Duchatel, J. P., Shivaprasad, H. L., Raidal, S. R., Abadie, J. X., Franciosini, M. P. & Smyth, J. A. (2008). Sequence comparison of pigeon circoviruses. Res Vet Sci 84, 311–319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.