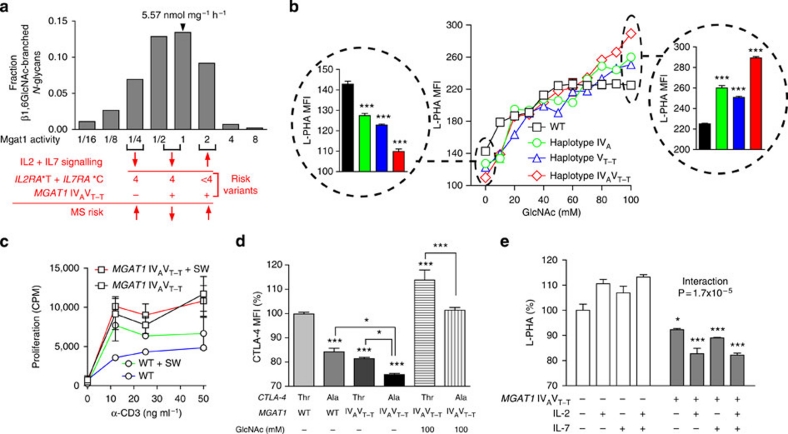
Figure 4. Conditional regulation of N-glycan branching and T cell growth by MGAT1 IVAVT−T.
(a) Ordinary differential equation model of medial Golgi enzyme reactions showing predicted alterations in β1,6GlcNAc branching when varying Mgat1 activity in the presence of fixed activities in Mgat2, 4 and 5. Activities in resting mouse 129/Sv T cells were used for Mgat1, Mgat2, Mgat5 (ref. 20), whereas Mgat4 was estimated. In red, predicted effects of IL2/IL7 signalling and various genetic combinations of IL2RA*T+IL7RA*C risk alleles (four alleles or less than four) on Mgat1 activity, β1,6GlcNAc branching and MS risk in the absence or presence of the MGAT1 IVAVT−T haplotype. (b) Mgat1-deficient Lec1 cells transiently transfected as indicated were co-incubated with or without GlcNAc for 3 days and then analysed by fluorescence-activated cell sorting (FACS) for L-PHA binding. P values by analysis of variance (ANOVA) and Newman–Keuls Multiple Comparison Test. (c) PBMCs heterozygous for the MGAT1 IVAVT−T haplotype (n=2 subjects assayed in triplicate) or homozygous for the common haplotype (WT; n=1 subject assayed in triplicate) were stimulated with anti-CD3 and analysed for proliferation as indicated for 2 days. Subjects used were IL2RA*T/C, IL7RA*CC and CTLA-4 Thr/Ala. Error bars represent range of all values. (d) Transiently transfected Lec1 cells with or without GlcNAc treatment (3 days) were analysed for CTLA-4 expression by fluorescence-activated cell sorting. P values by ANOVA and Newman–Keuls Multiple Comparison Test. (e) PBMCs with the indicated genotype (n=2) were stimulated with anti-CD3 (200 ng ml−1) and treated with IL-2 (20 ng ml−1) and IL-7 (20 ng ml−1) for 5 days. Subjects used were IL2RA*T/C, IL7RA*CC and CTLA-4 Thr/Thr. Data is the average of triplicate values for each subject. P values by 2-way ANOVA and Bonferoni's multiple comparison test. *P<0.05, **P<0.01, ***P<0.001 in all panels. Error bars are standard error of triplicate or greater values in all panels unless otherwise stated.