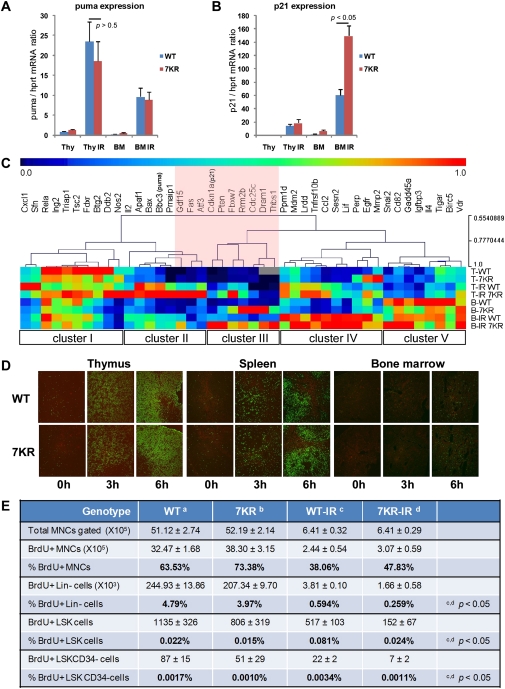
Figure 6.
Differential gene expression and cell cycle kinetics in p537KR mice. (A) The p53-inducible proapoptotic gene puma was expressed at higher levels in thymus than in BM after irradiation. The level of puma expression was similar between WT and p537KR mice. (B) The p53-inducible cell cycle arrest gene p21 was highly expressed in BM after irradiation, and showed even greater expression in p537KR animals. (A,B) Error bars represent the SEM from three animals. (C) Expression of 43 p53 target genes from thymus (T) and BM (B) prior to irradiation or 3 h post-irradiation (IR) was detected using microfluidic qPCR analysis. Relative expressions of each gene from all samples were scaled from 0 to 1, followed by hierarchical clustering analysis. (Cluster I) Repressive or less responsive genes. (Cluster II) Highly IR-induced genes in thymus. (Cluster III) Genes highly affected by 7KR mutations in BM. (Cluster IV) Highly IR-induced genes in BM. (Cluster V) Highly expressed genes in BM. The analysis suggests that several genes (highlighted in pink) are differentially affected by the 7KR mutations. (D) Thymi, spleens, and BM were isolated from mice at 0, 3, and 6 h after 5 Gy of whole-body irradiation. Apoptosis was detected by TUNEL staining (green). (E) Nonirradiated mice or mice exposed to 5 Gy of irradiation were injected with 2 mg of BrdU followed by 2-d administration of BrdU water (1 mg/mL). BMs were then isolated and stained with antibodies against lineage (Lin) markers; cell surface markers Sca1, c-kit, and CD34; and BrdU, followed by flow analysis. BrdU incorporated cell in Lin−, LSK (Lin−Sca1+c-kit+) and LSKCD34− subcell populations were analyzed. Error bars represent the SD from three animals.