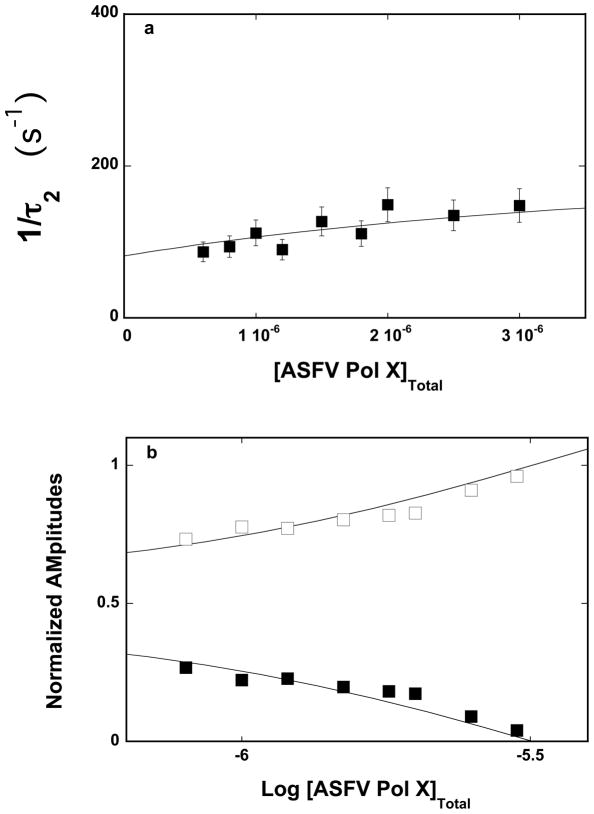
Figure 2.
a. The dependence of the reciprocal of the relaxation time, 1/τ2, for the binding of the ssDNA 10-mer, dεA(pεA)9, to the strong DNA-binding subsite of the ASFV pol X in buffer C (pH 7.0, 10°C), upon the total concentration of enzyme. The solid line is the nonlinear least-squares fit according to the two-step sequential mechanism defined by eq. 7, with the rate constants included in Table 1 (details in text). The error bars are standard deviations obtained from 3 – 4 independent experiments. b. The dependence of the normalized, individual relaxation amplitudes of the corresponding relaxation processes, A1 and A2, upon the logarithm of the total concentration of the polymerase. The solid lines are nonlinear least-squares fits, according to the two-step sequential mechanism, defined by eq. 7, with the relative fluorescence intensities included in Table 1. The rate constants are the same as obtained from the relaxation time analysis (Table 1); A1 (❑), A2 (■).