Summary
Mycoplasma pneumoniae is a wall-less human respiratory tract pathogen that colonizes mucosal epithelium via a polar terminal organelle having a central electron-dense core and adhesin-related proteins clustered at a terminal button. A mutant lacking J-domain co-chaperone TopJ is noncytadherent and nonmotile, despite having a core and normal levels of the major cytadherence-associated proteins. J-domain co-chaperones work with DnaK to catalyze polypeptide binding and subsequent protein folding. Here we compared features of the topJ mutant with other cytadherence mutants to elucidate the contribution of TopJ to cytadherence function. The topJ mutant was similar ultrastructurally to a non-cytadherent mutant lacking terminal organelle proteins B/C, including aberrant core positioning and cell morphology in thin sections, but exhibited a hybrid satellite growth pattern with features of mutants both having and lacking a core. Time-lapse images of mycoplasmas expressing a YFP fusion with terminal organelle protein P41 suggested that terminal organelle formation/positioning was delayed or poorly coordinated with cell growth in the absence of TopJ. TopJ required a core for localization, perhaps involving HMW1. P1 trypsin accessibility on other non-cytadherent mutants was significantly enhanced over wild-type but unexpectedly was reduced with topJ mutant cells, suggesting impaired processing, translocation, and / or folding of this adhesin.
Keywords: J-domain, co-chaperone, cytoskeleton, cytadherence, terminal organelle
Introduction
The cell wall-less bacterial pathogen Mycoplasma pneumoniae is a leading cause of community-acquired pharyngitis, bronchitis, and atypical or “walking” pneumonia (Waites and Talkington, 2004). Both gliding motility and binding to host cell receptors (cytadherence) contribute to evasion of mucociliary clearance and successful colonization of host mucosal epithelium (Jordan et al., 2007; Lipman and Clyde, 1969). Gliding and cytadherence are mediated by the mycoplasma terminal organelle, a membrane-bound cell extension defined by its complex electron-dense core and adhesin proteins clustered at its distal end (Bredt, 1968; Krause and Balish, 2004; Biberfeld and Biberfeld, 1970). Assembly of a functional terminal organelle involves the coordinated incorporation of cytoskeletal components that appear to stabilize the final structure (Krause and Balish, 2004).
Localization of the 170-kDa adhesin P1 to the terminal organelle is necessary but not sufficient for cytadherence competence. For example, loss of cytoskeletal proteins HMW1 or HMW2 results in failure to cluster P1 at a cell pole and the inability to cytadhere (Baseman et al., 1982), whereas loss of the adhesin-related P30 or cytadherence-associated proteins B and C results in the inability to cytadhere (Krause et al, 1982) despite apparently normal P1 trafficking (Seto and Miyata, 2003; see Table 1 for summary of mutants). Membrane proteins B and C, like P1, localize primarily to the distal end of the terminal organelle (Dallo et al., 1990; Feldner et al., 1982; Franzoso et al., 1993; Layh-Schmitt and Herrmann, 1992; Seto et al., 2001) and with cytoskeletal proteins HMW1, HMW3, and P65, cross-link with P1 in protein proximity studies (Layh-Schmitt and Herrmann, 1994; Layh-Schmitt et al., 2000). Proteins B and/or C secure P1 to the Triton X-100-insoluble cytoskeleton fraction [triton shell; (Layh-Schmitt and Harkenthall, 1999; Gobel et al., 1981; Meng & Pfister, 1980)], and P1-B complexes can be purified from wild-type M. pneumoniae cells in the absence of chemical crosslinking (Nakane et al., 2011). Membrane protein P30 is likewise required for P1 function and cross-links with P1 in protein proximity studies (Layh-Schmitt et al., 2000; Romero-Arroyo et al., 1999), but unlike proteins B, C, and P1, P30 localizes exclusively to the distal end of the terminal organelle (Seto et al., 2001). Epitope mapping and adherence inhibition analyses suggest that functional maturation and receptor-binding competence requires proper folding of three discontinuous regions of P1 to within close proximity (Gerstenecker and Jacobs, 1990).
Table 1.
Summary of the protein profiles of wild-type M. pneumoniae, recombinant topJ derivatives, and several terminal organelle mutants.
| Strain | Proteina | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| P1 | B/C | HMW1 | HMW2 | P30 | P24 | TopJ | HA b | Reference | |
| Wild-type | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | Krause et al., 1982 |
| topJ | +++ | +++ | +++ | +++ | +++ | + | − | − | Cloward and Krause, 2009 |
| P24− | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | Hasselbring and Krause, 2007b |
| I-2 | +++ | +++ | + | − | ++ | NTc | +++ | − | Krause et al., 1982 |
| II-3 | +++ | +++ | +++ | +++ | − | NT | +++ | − | Krause et al., 1982 |
| III-4 | +++ | − | +++ | +++ | +++ | NT | +++ | − | Krause et al., 1982 |
| M6 | +++ | +++ | − | + | Δ++d | NT | +++ | − | Layh-Schmitt et al., 1995 |
| H9 | +++ | +++ | + | Δ++ | ++ | NT | +++ | − | Hedreyda and Krause, 1995 |
| topJΔHPD | +++ | +++ | +++ | +++ | +++ | +++ | +++e | − | Cloward and Krause, 2010 |
| topJΔC | +++ | +++ | +++ | +++ | +++ | + | Δ+ | +++ | Cloward and Krause, 2010 |
| topJΔAPR | +++ | +++ | +++ | +++ | +++ | + | Δ+ | ++++ | Cloward and Krause, 2010 |
Relative amounts of indicated protein; − (none) to +++ (wild type)
Hemadsorption capacity
Not tested
Truncated proteins designated with Δ
Wild-type levels of the TopJ derivative protein TopJΔHPD
Given the complexity of protein interactions required for terminal organelle assembly and maturation, the identification of the chaperone DnaK as a component of the triton shell in close proximity to P1 is not surprising (Regula et al., 2001; Layh-Schmitt et al., 2000). DnaK functions in polypeptide folding, protein translocation, and macromolecular assembly, requiring a J-domain co-chaperone and the nucleotide exchange factor GrpE for peptide binding and release (reviewed in Frydman, 2001; Hartl and Hayer-Hartl, 2002). The J-domain co-chaperone TopJ of M. pneumoniae localizes to the terminal organelle and is required for cytadherence and gliding motility, despite wild-type levels of P1, P30, and all other cytadherence-related proteins examined (Cloward and Krause, 2009). P1, HMW1, P30, and P65 appear to co-localize normally in the topJ mutant, but cellular positioning of the terminal organelle is defective, suggesting a likely role for TopJ, and by inference DnaK, in terminal organelle maturation (Cloward and Krause, 2009). Complementation of the topJ mutant with recombinant TopJ, but not a derivative having an altered canonical HPD domain, rescues cytadherence and gliding, underscoring the importance of co-chaperone function in the topJ mutant phenotype (Cloward and Krause, 2009; Cloward and Krause, 2010).
Here we compared the topJ and other cytadherence mutants to elucidate more specifically how TopJ function contributes to terminal organelle maturation in the context of other terminal organelle components. Core positioning and both size and electron density of cells, as viewed in thin sections, differed among the mutants, underscoring the varied influences of terminal organelle proteins on cellular architecture. By this analysis, the topJ mutant most closely resembled mutant III-4, which lacks cytadherence-associated proteins B and C. Satellite growth, immunofluorescence analysis, and cell growth patterns suggested that TopJ requires an electron-dense core for polar localization and contributes to terminal organelle maturation, migration, or both during cell division. Finally, accessibility of adhesin P1 to trypsin was reduced with the topJ mutant relative to wild-type M. pneumoniae and other cytadherence mutants, suggesting that P1 fails to translocate and/or fold correctly on the mycoplasma cell surface in the absence of this co-chaperone, perhaps impairing the capacity of a P1/B/C/P30 complex to anchor the terminal organelle core properly to the mycoplasma membrane.
Results
Core positioning in topJ and other non-cytadhering mutants
We previously noted that terminal organelle core positioning is defective in M. pneumoniae topJ mutant cells (Cloward and Krause, 2010) and expanded here the analysis of core placement compared to other terminal organelle mutants by categorizing the extent of core extension beyond the cell body and determining the percentages observed for each category (Figure 1). The categories were: fully extended cores (100%); cores perpendicular to the membrane and extended from the cell body approximately 75%, 50%, or 25%; cores parallel to the membrane (Henderson and Jensen, 2006) and categorized here 50%*, and cores perpendicular to the membrane but unextended (0%). Wild-type M. pneumoniae and the P24− mutant had the highest percentage of cells (ca. 65– 70%) with fully extended cores (Figure 1), indicating that the reduced steady-state level of P24 in the topJ mutant (Cloward and Krause, 2009) was not responsible for defective core positioning. The remaining mutants had higher percentages of cores not fully extended, with the topJ and III-4 mutants having the largest combined percentages of cells with partially-extended, perpendicularly-oriented cores (ca. 36 and 27%, respectively). As expected (Cloward and Krause, 2010), the topJ deletion derivative topJΔHPD resembled the topJ mutant, while topJΔC and topJΔAPR were more similar to wild-type. Curiously, the electron-lucent space flanking each core was retained regardless of core positioning, suggesting the presence of a poorly staining component that excludes cytoplasmic content.
Figure 1. Electron-dense core positioning in wild-type M. pneumoniae and several terminal organelle mutants.
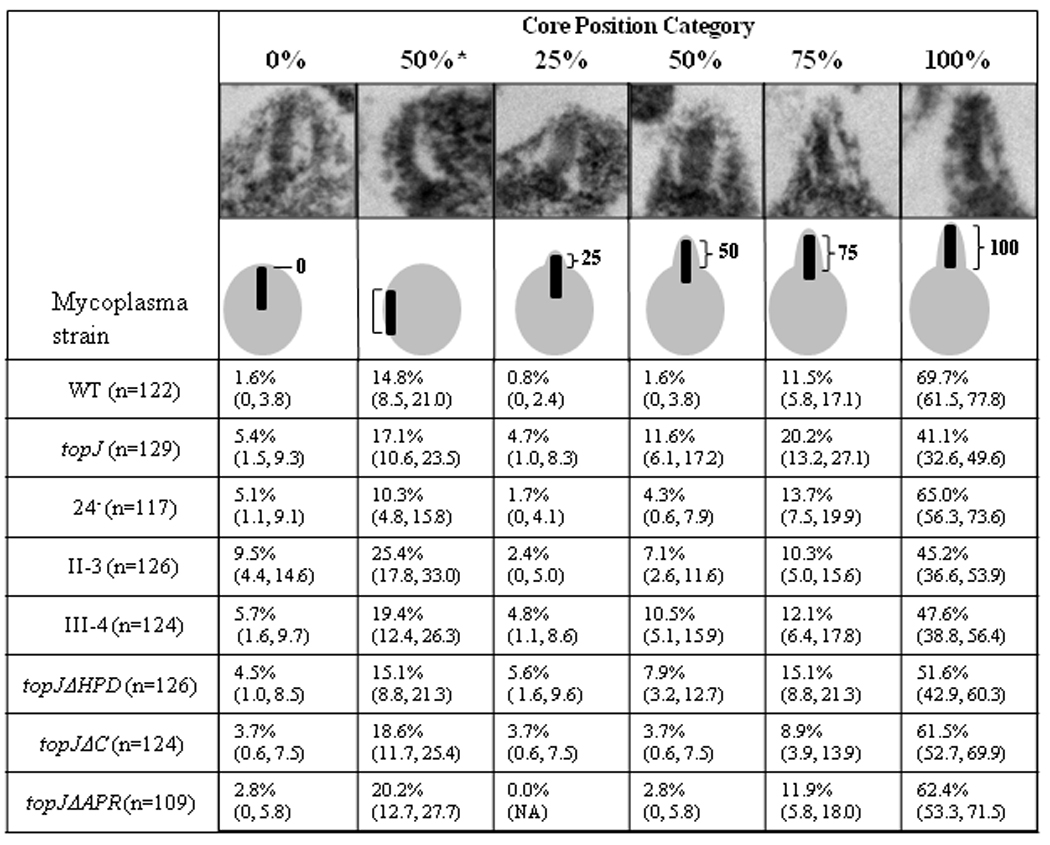
Core position categories were established based on the length of core extension beyond the cell body / total core length. A representative TEM image and schematic depiction are provided for each category. Values are indicated as percentages observed, followed by the percentage range at a 95% confidence interval (minimum, maximum), N = total cells examined per strain.
Ultrastructural appearance of topJ and other non-cytadhering mutants
M. pneumoniae cells with no TopJ or with the topJΔHPD derivative exhibit fragility under a variety of fixation conditions, resulting in an enlarged, electron-lucent appearance in thin section (Cloward and Krause, 2010). Here we examined additional mycoplasma mutants for a possible correlation with the topJ mutant phenotype (Figure 2; Table 2). Cell sizes and electron densities in thin sections were quantified and sorted, with size values in the top 25% for each assumed to represent mid-cell sections and thus to reflect relative cell size most accurately (Table 2). Wild-type and P24− mutant cells in thin section were equivalent in size but significantly smaller than mutant II-3, III-4, or topJ cells (p ≤ 0.01); mutant III-4 dwarfed all strains in both average and maximum areas (p ≤ 0.05 compared with either mutant II-3 or the topJ mutant, and p ≤ 0.001 compared with the M6 or P24- mutants or wild-type cells). Noncytadherent mutants grow in suspension in liquid culture, but the topJ and II-3 mutant cell sizes in thin section surpassed M6 by ≥30% (p ≤ 0.01), suggesting that failure to attach to a solid surface during growth was not solely responsible for the size difference. Although topJ and II-3 mutant cells were almost identical and statistically indistinguishable in size, the topJ mutant most closely resembled mutant III-4 in overall electron density (Table 2). We also standardized for cell size by examining cell densities in thin section for each mutant population only within the range of larger wild-type cells (0.07–0.21 µm2), but both the III-4 and topJ mutants continued to exhibit the lowest overall electron densities (Table 2; p values ranged from ≤ 0.001 to ≤ 0.05, see Supplemental Table 1). Their reduced cell densities suggest that these cells resisted staining and lost cell content during processing for electron microscopy. Finally, both the III-4 and topJ mutants exhibited fibrous strands extending outward from apparent membrane disruptions, suggesting membrane fragility (data not shown).
Figure 2. Ultrastructural comparisons of wild-type M. pneumoniae and terminal organelle mutants.
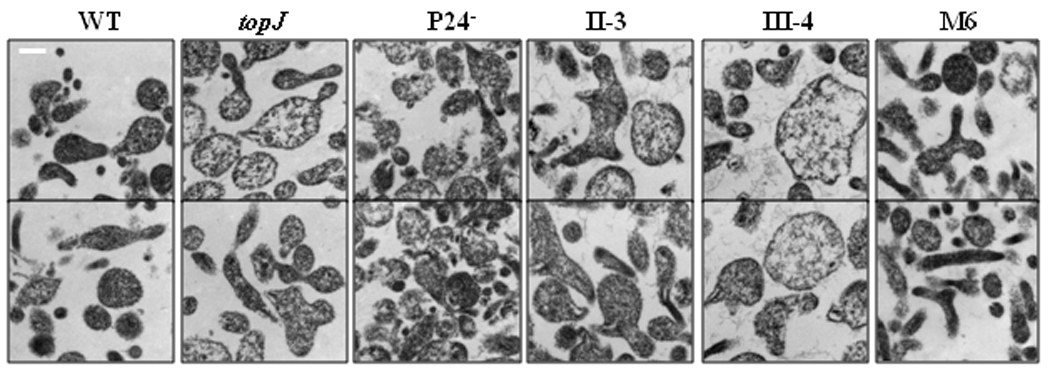
Representative TEM micrographs of M. pneumoniae strains as indicated. Bar, 200 nm.
Table 2.
Ultrastructural analysis of M. pneumoniae topJ deletion derivatives.
| wild-type a | topJa | P24− | II-3 | III-4 | M6 | |
|---|---|---|---|---|---|---|
| Total cells - number | 221 | 207 | 201 | 208 | 206 | 213 |
| Mean section area in µm2 (% wild-type) | 0.072 (100) | 0.119 (165) | 0.085 (117) | 0.115 (159) | 0.182 (250) | 0.093 (127) |
| Median section area in µm2 (% wild-type) | 0.066 (100) | 0.086 (131) | 0.076 (115) | 0.088 (133) | 0.132 (199) | 0.069 (105) |
| Top 25% - numberb | 56 | 52 | 51 | 51 | 54 | 54 |
| Mean section area in µm2 (% wild-type) | 0.142 (100) | 0.273 (192) | 0.156 (109) | 0.256 (180) | 0.413 (290) | 0.197 (139) |
| Median section area in µm2 (% wild-type) | 0.134 (100) | 0.220 (165) | 0.140 (105) | 0.228 (171) | 0.343 (257) | 0.171 (128) |
| Range in µm2 | 0.107–0.291 | 0.152–0.692 | 0.110–0.317 | 0.152–0.542 | 0.227–0.897 | 0.119–0.437 |
| Electron density as % wild-type | ||||||
| Total population | 100 | 81 | 97 | 98 | 83 | 105 |
| Within cell section range b 0.70–0.21 µm2 | 100 | 80 | 96 | 98 | 86 | 103 |
See Supplemental Tables 1 and 2 for statistical comparisons
Cell growth and TopJ localization patterns differed based on core presence
P1 clusters primarily at the distal end of the terminal organelle in wild-type M. pneumoniae, with TopJ localizing proximally (Figure 3; Baseman et al., 1982; Feldner et al., 1982; Cloward and Krause, 2009). P1 likewise forms a focus at the apex of cell branches and occasionally at a cell pole in topJ and core-positive mutants II-3 and III-4 (Romero-Arroyo et al., 1999; Seto and Miyata, 2003; Cloward and Krause, 2009; Cloward and Krause, 2010), but core-negative mutants I-2 and M6 exhibit a diffuse and punctate rather than focal P1 distribution (Baseman et al., 1982; Seto et al., 2001; Seto and Miyata, 2003). Here we examined TopJ localization relative to P1 in core-positive and core-negative mutants (Figure 3). TopJ co-localized proximal to P1 in core-positive mutants II-3 and III-4 at either a cell pole or the apex of a branched cell, likely corresponding to a terminal organelle (Figure 3, white circles). Interestingly, some separation was noted between P1 and TopJ in mutant III-4 but not in other mutants, but we have not attempted to quantify this observation. Like P1, TopJ exhibited a diffuse and punctate distribution in core-negative mutant M6, with no apparent correspondence in the fluorescence patterns for each. In contrast, TopJ fluorescence was typically more focal in core-negative mutant H9, with clear correspondence between some (white circles) but not all (red and yellow circles) P1 and TopJ fluorescent foci. Furthermore, some polar foci were clearly noted with mutant H9 (arrows). The distinct fluorescence patterns for M6 and H9 may simply reflect differences in morphology between the two mutants, but attempts to otherwise quantify these differences were hampered by the inability to define individual cells for each. Nevertheless, taken together, these data suggest that an intact core, and perhaps more specifically HMW1, which is absent in mutant M6 but only reduced in mutant H9, are required for TopJ localization.
Figure 3. Immunofluorescence microscopy analysis of TopJ localization in wild-type M. pneumoniae and cytadherence mutants.
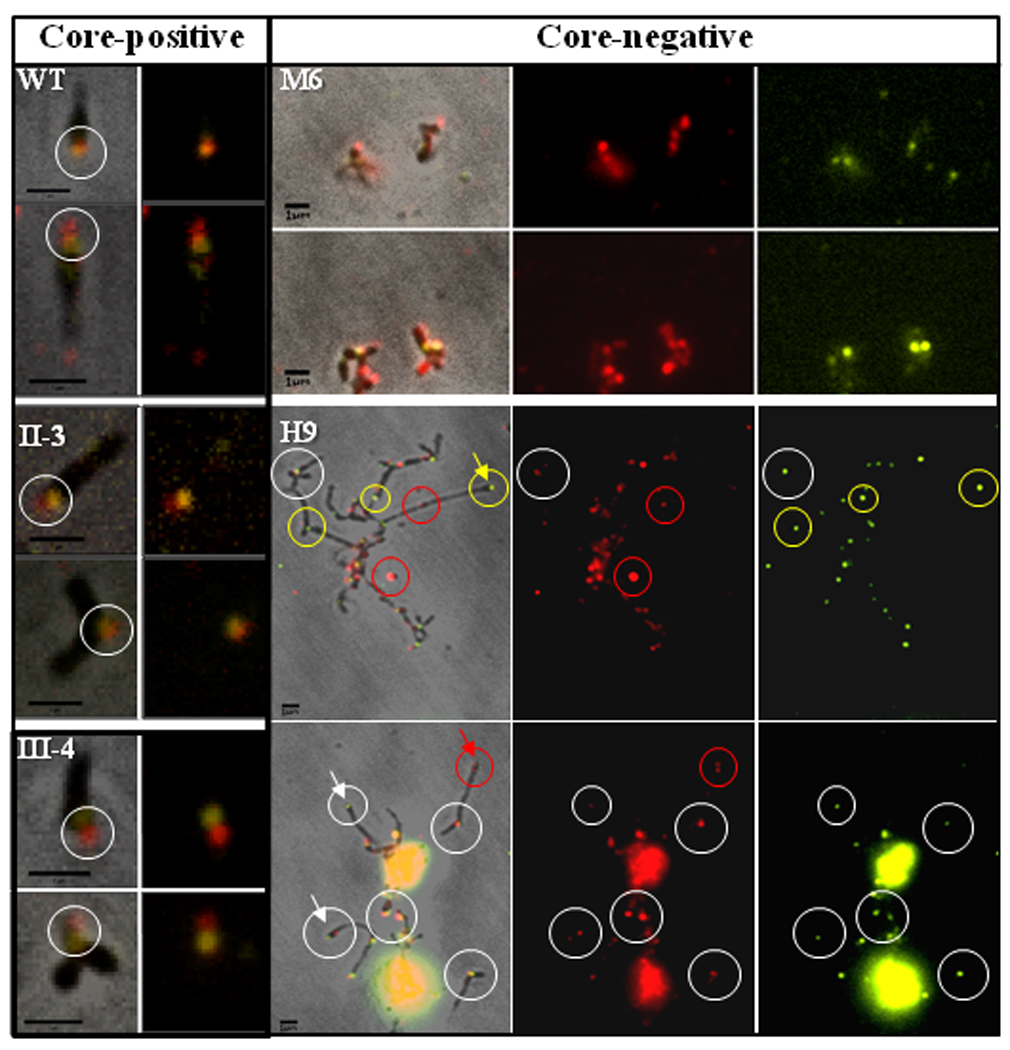
P1 (false-colored red) and TopJ (false-colored yellow) were immunolocalized in wild-type (WT) and mutant strains as indicated. For WT, II-3, and III-4: left panel, merged phase contrast and fluorescence images; right panel, fluorescence images only. White circles indicate co-localization of TopJ and P1. For M6 and H9: left panel, merged phase contrast and fluorescence images; middle panel, P1; right panel, TopJ. White circles, representative examples of co-localized P1 and TopJ foci; red and yellow circles, representative P1 foci lacking clear, corresponding Top J foci and vice versa, respectively; arrows, polar foci for P1 and Top J (white) and TopJ only (yellow) in mutant H9. Bar, 1µm.
Delayed terminal organelle formation and/or migration in topJ mutant cells
To compare topJ and other cytadherence mutants further we analyzed their colony growth and terminal organelle development patterns (Figure 4). In contrast to the outward spreading and satellite growth of wild-type M. pneumoniae, the core-positive mutants II-3 and III-4 formed isolated microcolonies that expanded with defined edges, while the core-negative mutants M6 and H9 exhibited outward growth characterized by cell branching and elongation (Figure 4A). The topJ mutant exhibited microcolony growth patterns characteristic of both core-positive and core-negative mutants, i.e. branched growth at the microcolony periphery like core-negative mutants, but also a pronounced center and defined edges like core-positive mutants. These mutants are all non-motile hence differences in microcolony formation are likely a function of cell growth and division. In order to follow cell growth over time for the topJ mutant we utilized a yellow fluorescent protein (YFP) fusion to terminal organelle protein P41 (Kenri et al., 2004) and time-lapse imaging (Figure 4B and 4C). As expected (Hasselbring and Krause, 2007a), wild-type cells (Figure 4B) formed a sharp YFP-P41 focus at each terminal organelle, including the leading end of gliding cells (arrows) and cells growing outward from a microcolony (arrowheads). However, YFP-P41 foci were typically absent as cell growth extended outward from microcolonies of the topJ mutant (Figure 4C, red circles). Rather, we observed diffuse fluorescence, with very faint foci forming in a manner that did not appear to be coordinated with cell growth (red arrowheads), consistent with the high incidence of cells with no evidence of an electron-dense core (Cloward and Krause, 2009).
Figure 4. Cell growth and terminal organelle development in wild-type and mutant M. pneumoniae.
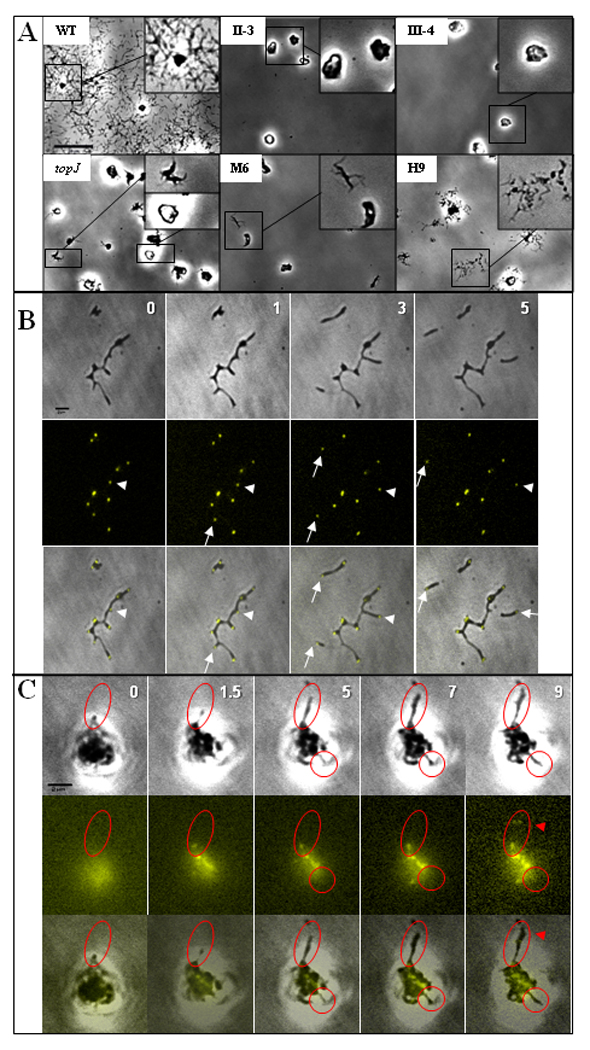
(A) Satellite growth analysis of wild-type (WT) M. pneumoniae cells and the indicated core-positive (topJ, II-3, III-4) and core-negative (M6, H9) cytadherence mutants. Mycoplasmas were incubated in SP4 medium with 3% gelatin for 72 hr. Bar, 20µm. (B and C) Time-lapse images of terminal organelle formation in wild-type (B) M. pneumoniae and the topJ mutant (C) expressing YFP-P41. Top row, phase contrast; middle row, YFP-P41 fluorescence false-colored yellow; bottom row, merged phase contrast and fluorescence images. Numbers, incubation time in hours; white arrows, terminal organelle on gliding cells; white arrowheads, terminal organelle on growing cells. Red circles, cells growing outward with no polar terminal organelle. Red arrowhead, faint YFP-P41 foci. Bar, 2µm.
Loss of TopJ alters protease accessibility to P1 adhesin
The topJ mutant has wild-type steady-state levels of all known cytadherence-associated proteins yet is non-cytadherent (Cloward and Krause, 2009), prompting us to examine accessibility of terminal organelle surface proteins to trypsin as a possible reflection of lack of adhesin function. Wild-type and topJ mutant cells were initially incubated with trypsin for 30 minutes and the digestion patterns compared (Figure 5A). As expected, steady-state levels of GroEL remained virtually unchanged in cells treated with trypsin alone, indicating that cell lysis was minimal during trypsinization, and that the increased fragility of the topJ and III-4 mutants with sample processing for electron microscopy was not evident here. As a control, full-length GroEL was barely detectable when cells were solubilized with Triton X-100 and trypsin- treated. Of the terminal organelle proteins examined, only P1 demonstrated a clear and reproducible difference, appearing less accessible to trypsin with the topJ mutant than with wild-type cells (data not shown and Figure 5A).
Figure 5. P1 accessibility to digestion by trypsin over time in wild-type and mutant strains.
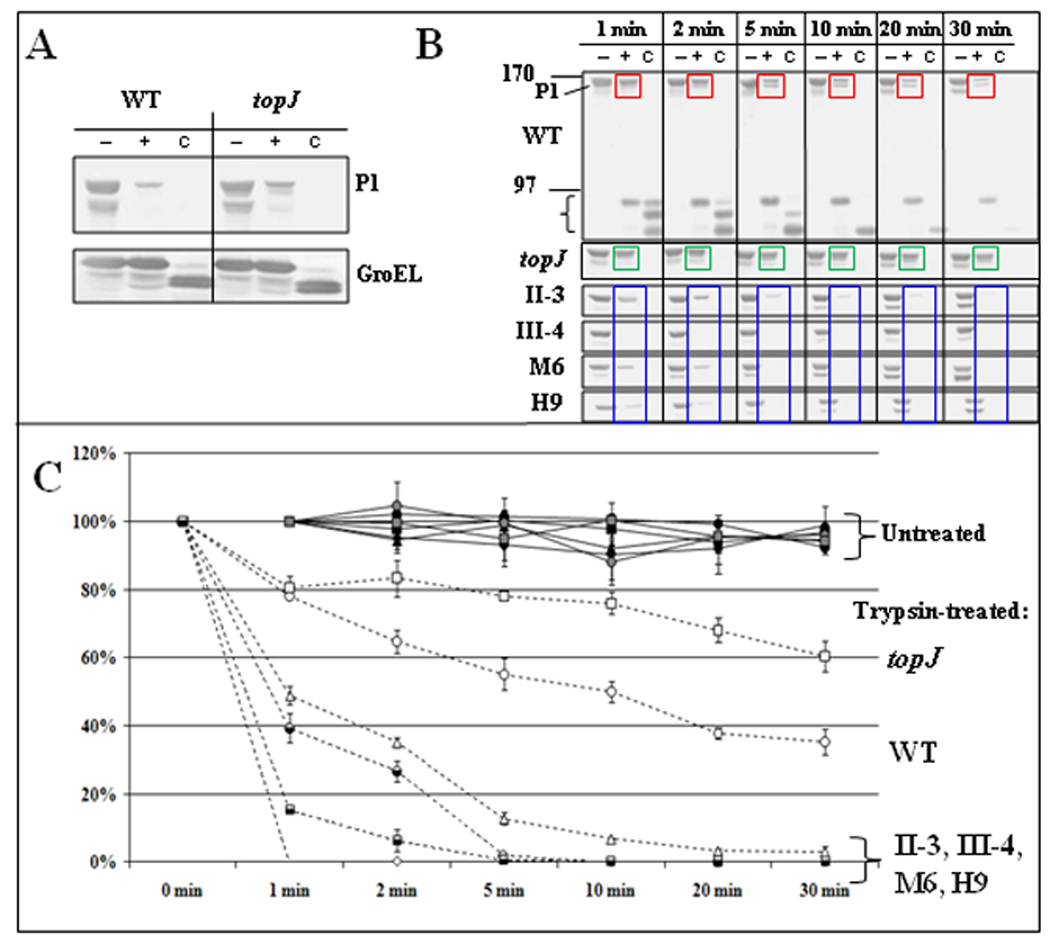
(A) Proof of concept. Immunoblot analysis of M. pneumoniae wild-type (WT) and topJ mutant cells after 30-min trypsin digestion and analysis by 4–12% gradient SDS-PAGE and immunoblotting with the antisera listed to right. (B) Representative immunoblots of wild-type (WT) and topJ mutant cells treated with trypsin for the indicated times, followed by SDS-PAGE and immunoblot analysis with P1-specific antiserum. The mutant blots are cropped to show only the reduction in the 170-kDa P1 band over time. −, no trypsin; +, trypsin-treated; C, samples solubilized with 2% Triton X-100 for 30 min at 37°C before trypsin treatment; bracket, P1 digestion products; colored boxes highlight changes in P1 over time with trypsinization of wild-type M. pneumoniae (red), the topJ mutant (green), and other cytadherence-mutants (blue). (C) The reduction in intensity of the 170-kDa P1 band for trypsin-treated and untreated samples over time. Bars, standard error of the mean. See Supplemental Table 3 for detailed statistical comparisons.
We expanded the analysis of P1 accessibility to protease through a quantitative time course with wild-type M. pneumoniae, the topJ mutant, and other non-cytadhering mutants (Figure 5B & 5C). Trypsin treatment of wild-type cells resulted in gradual loss of the 170-kDa P1 band (Figure 5B, red boxes) and coincident appearance of < 90-kDa P1 products, which then diminished over time (bracket). The relative intensity of the P1 bands in untreated wild-type and topJ mutant cells decreased by <10% over 30 min at 37°C (Figure 5C). In order to obtain the percentage of the P1 band remaining and normalize between Western blots, the band intensities of trypsinized P1 at each time point were measured relative to band intensities of untreated P1 at the same time point (Figure 5C). P1 levels from trypsin-treated wild-type cells decreased by about 20% and 60% at 1 and 30 min, respectively (Figure 5C). P1 from topJ mutant cells also decreased by about 20% after 1 min but exhibited a slower decline than wild-type P1 over 30-min, to a final reduction of 40% [Figure 5B, green boxes, and Figure 5C; p ≤ 0.01 at 5, 10, 20 min; p ≤ 0.05 at 30 min (see Supplemental Table 2 for all p-values)]. Surprisingly, P1 digestion was strikingly faster for mutants II-3, III-4, M6, and H9 (60%–100% reduction within 2 min), with full-length P1 almost completely absent in these mutants by 30 min (Figure 5B, blue boxes, and Figure 5C; p ≤ 0.01). Thus, P1 was significantly more trypsin-accessible in both core-positive and core-negative cytadherence mutants than in wild-type, but conversely was less trypsin-accessible in the absence of TopJ than for all other strains examined. We likewise assessed topJ deletion derivatives (Cloward and Krause, 2010) for P1 accessibility to trypsin (Figure 6). The topJ and topJΔHPD cells exhibited similar and statistically indistinguishable P1 digestion patterns, linking altered P1 trypsin accessibility directly to TopJ co-chaperone function. In contrast, the pattern for topJΔC cells more closely aligned with and was statistically indistinguishable from that of wild-type over the time course, while the topJΔAPR derivative had an intermediate rate of P1 digestion that was statistically indistinguishable from either the topJ mutant or wild-type after 30 min. Interestingly, the loss of TopJ did not affect Triton-partitioning of all terminal organelle proteins examined, nor did the loss of these terminal organelle proteins affect TopJ partitioning (data not shown), suggesting that association with the triton shell is protein-specific, TopJ-independent, or both.
Figure 6. P1 accessibility to digestion by trypsin over time for topJ deletion derivatives.
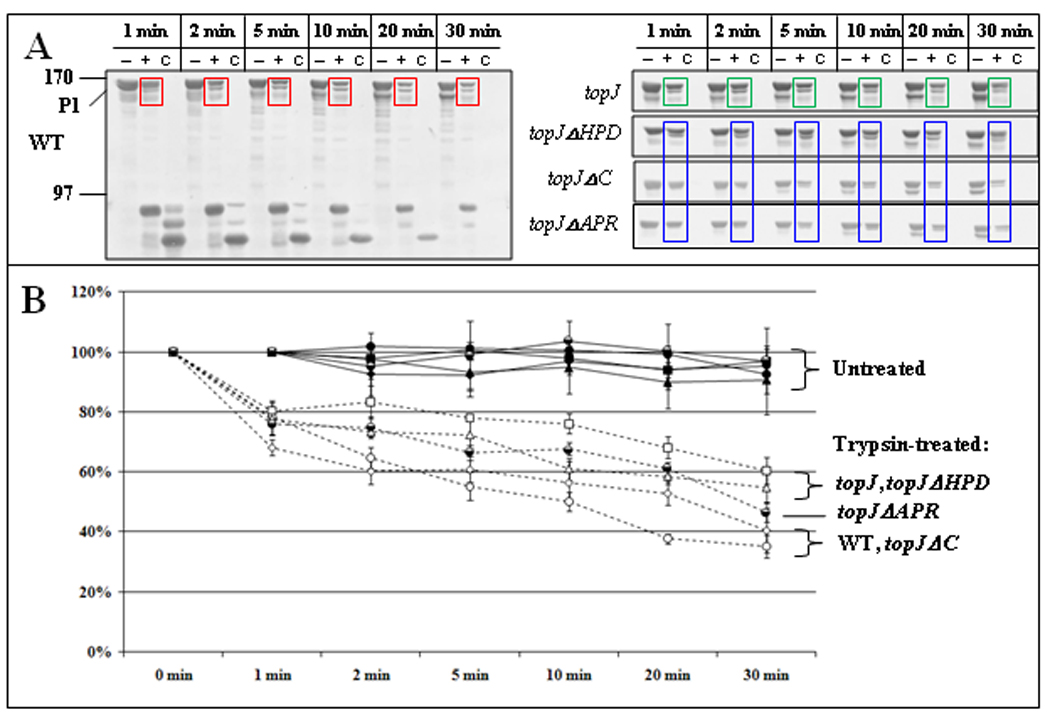
(A) Representative immunoblots of the indicated strains treated with trypsin for the indicated times followed by separation by SDS-PAGE and Western immunoblot analysis, as described for figure 5. Colored boxes highlight changes in P1 over time with trypsinization of wild-type M. pneumoniae (red), the topJ mutant (green), and the indicated topJ deletion derivatives (blue) (B) The reduction in intensity of the 170-kDa P1 band for trypsin-treated and untreated samples over time. Bars, standard error of the mean. See Supplemental Table 3 for detailed statistical comparisons.
Discussion
Cytadherence competence in M. pneumoniae requires the trafficking of P1 to the terminal organelle, where it appears to form an adhesin complex with B, C, and P30 (Hahn et al., 1998; Layh-Schmitt and Harkenthal, 1999; Layh-Schmitt and Herrmann, 1994; Nakane et al., 2011) and likely undergoes final folding to assume a functional conformation for receptor binding. The topJ mutant has wild-type levels of known cytadherence-associated proteins, which co-localize focally with terminal organelle core components yet fail to function in adherence and gliding (Cloward and Krause, 2009; Cloward and Krause, 2010). Here we explored further the topJ mutant phenotype in the context of other terminal organelle mutants.
Henderson and Jensen (2006) previously described electron-dense cores that fail to extend from the cell body in wild-type M. pneumoniae, and our analysis here quantified their prevalence in wild-type and mutant cell populations. We believe that two major conclusions emerge from analysis of core positioning data in Figure 1. First, >80% of cores examined in wild-type cells were extended by 75% or more, compared to approximately 55–60% for the topJ, II-3, and III-4 mutants, with no overlap at the 95% confidence intervals at 100% extension. This difference suggests a defect in core positioning in the absence of P30 or B & C, both of which likely partner with the major adhesin P1 in the terminal organelle membrane (Layh-Schmitt and Harkenthall, 1999; Nakane et al., 2011). Similarly defective core positioning in the absence of TopJ raises the possibility that this co-chaperone is required for formation of a functional P1/B/C/P30 adhesin complex. Second, core placement in the topJ mutant was most like that of topJΔHPD, indicating that the canonical J-domain, and by inference DnaK, are required for normal core positioning. While core-placement patterns varied among the mutant strains, suggesting that the corresponding terminal organelle proteins influence core positioning differently, a considerably larger sample size is required to make relevant conclusions regarding those mutants.
The topJ and III-4 mutants were similar with respect to the size, electron density, and integrity of cells in thin sections, suggesting that sample processing for EM was consistently less effective for these mutants than for wild-type cells or the II-3, M6, or P24− mutants. EM analysis of the M. pneumoniae triton-insoluble fraction reveals membranous material associated with the triton shell (Regula et al., 2001). Unlike the triton-soluble P30, proteins B, P1, and TopJ partition with both the insoluble and soluble fractions (data not shown) and may form higher-ordered complexes between the membrane and cytoskeleton, perhaps thereby contributing to cell integrity. Curiously, mutant M6 cells had a normal electron density despite the loss of HMW1, a predominantly triton-insoluble peripheral membrane protein critical for electron-dense core formation (Balish et al., 2001; Seto and Miyata, 2003), suggesting that HMW1 may contribute to cell integrity differently than P1, B/C, or TopJ.
The topJ mutant exhibited a hybrid satellite growth pattern, with features typical of both core-positive and core-negative cytadherence mutants, and terminal organelle protein P41 was not necessarily associated with extending filaments of topJ mutant cells, unlike wild-type M. pneumoniae. In addition, in this and previous studies we showed that individual cells of the topJ mutant lack paired foci for several terminal organelle proteins (Cloward and Krause, 2009; Hasselbring et al., 2006; Seto at al., 2001; Seto and Miyata, 2003). Finally, TopJ appeared more focal in mutant H9 than in mutant M6, often paired with P1, suggesting that reduced HMW1 in H9 partially supports TopJ localization compared to M6, which lacks HMW1 entirely. These data are consistent with our previous finding that HMW1 is required for P1 trafficking (Hahn, et al., 1998), and a scenario where P1 fails to interact normally with P30, B and C in the absence of TopJ, impacting both the core interface with the terminal organelle membrane as well as functional maturation of the adhesin complex. While the basis for absence of a terminal organelle in some cells of the topJ mutant remains unclear, the potential influence of reduced P24 on terminal organelle formation is intriguing. The absence of P24 does not affect core positioning or cell integrity (this study) but does impact temporal and spatial patterns for new terminal organelle formation (Hasselbring and Krause, 2007b), albeit in a manner distinct from the striking YFP-P41 patterns seen here in growing topJ mutant cells (Figure 4C), and which is likely not due simply to the reduced levels of P24 in the absence of TopJ. However, the APR and C-terminal domains of TopJ are required for P24 stabilization (Cloward and Krause, 2010) and perhaps for normal core-positioning (Figure 1). Additional studies are clearly required to define how P24 influences the timing and location of new terminal organelle formation and how this process is linked to co-chaperone function.
Proper P1 trafficking alone is not sufficient for adherence competence. To explore P1 maturation specifically, we examined its accessibility to trypsin treatment of wild-type and mutant whole cells. In the absence of proteins P30, B/C, HMW1, or HMW2, proteolysis of P1 was dramatically accelerated relative to that in wild-type cells. Nearest-neighbor analyses place these proteins in close proximity to P1 (Layh-Schmitt and Herrmann, 1994; Layh-Schmitt et al., 2000), and their absence appears to increase accessibility to trypsin, as reflected in their rapid digestion. However, conformational influences in addition to shielding could account for accelerated P1 proteolysis, as supported by the accelerated proteolysis of P1 in mutants having B/C and P30 but lacking a core. In striking contrast, P1 accessibility to trypsin was significantly reduced in the topJ mutant relative to wild-type, suggesting that in the absence of TopJ, P1 achieves a trypsin-resistant conformation, fails to translocate to the surface at wild-type levels, or both. While either scenario might impact the interaction of P1 with B/C and P30, the clear difference in accessibility of P1 to trypsin indicates that TopJ impacts P1 maturation in a manner distinct from that of accessory proteins P30 and B/C. In addition, the similar P1 digestion timecourse for the topJ mutant and topJΔHPD deletion derivative indicates the essential role of TopJ co-chaperone function specifically in terminal organelle maturation. Nakane et al. (2011) identified two major trypsin sites for P1, one of which coincides with a region targeted by adherence-inhibiting antibodies. This finding adds functional relevance to the correlation seen here between differences in P1 trypsin accessibility between wild-type M. pneumoniae and cytadherence mutants.
Previously, TopJ was defined as a J-domain co-chaperone, likely interacting with DnaK for terminal organelle protein maturation (Cloward and Krause, 2010). J-domain proteins activate the ATPase of DnaK, catalyzing protein binding and subsequent folding and / or translocation (Hendrick and Hartl, 1995). In the absence of a J-domain protein, DnaK can continue to act alone but less efficiently (Liberek et al., 1991), likely resulting in the misfolding of proteins. Furthermore, DnaK can be membrane-associated in Escherichia coli, possibly to facilitate protein translocation (Bukau et al., 1993). In M. pneumoniae, DnaK partitions in the triton shell and localizes in close proximity to P1 (Layh-Schmitt et al., 2000; Regula et al., 2001); therefore, loss of co-chaperone TopJ may limit DnaK activation, resulting in a cascade of morphological and functional manifestations. While other scenarios are possible, we propose that TopJ associates with HMW1, which is required for trafficking of P1 to the terminal organelle, and interacts with DnaK to facilitate folding, processing, and / or translocation to the surface to achieve a functional conformation for P1, B/C, or perhaps both.
Experimental Procedures
Bacterial strains and growth conditions
Wild-type M. pneumoniae strain M129-B18 (Lipman and Clyde, 1969), mutant II-3, mutant III-4 (Krause et al., 1982) mutant M6 (Layh-Schmitt et al., 1995), mutant H9 (Hedreyda and Krause, 1995), mutant P24− (Hasselbring and Krause, 2007a, 2007b), the topJ mutant (Cloward and Krause, 2009), and topJ deletion derivatives (Cloward and Krause, 2010) were grown in tissue culture flasks with SP4 medium (Tully et al., 1977) at 37°C until mid-log phase. The H9 and topJ mutants were grown with gentamicin (18 µg/ml); P24− and the topJ deletion derivatives were grown with gentamicin and chloramphenicol (24 µg/ml). For terminal organelle development studies, wild-type M. pneumoniae and topJ mutant cells were each transformed with yfp-p41 in Tn4001cat conferring chloramphenicol resistance (Hedreyda et al., 1993; Kenri et al., 2004), expanded in SP4 medium with appropriate antibiotics, and screened for YFP-P41 (data not shown).
Microscopy
Sample preparation for TEM and digital image acquisition of thin sections was performed as described previously (Willby and Krause, 2002). Mycoplasmas were prepared and examined for immunofluorescence microscopy as described (Cloward and Krause, 2009), or with modifications to yield more individual cells. Briefly, overnight cultures were harvested, syringe-passaged, filtered (0.45 µm), and incubated in fresh growth medium 2 h at 37°C prior to fixation. Phase contrast / fluorescence microcinematographic analysis of terminal organelle development was performed as described (Hasselbring et al., 2005) except that cultures were incubated overnight, after which the medium was exchanged with fresh SP4 + 3% gelatin and appropriate antibiotics (pH 7.2) and maintained at 37°C for 30 min prior to viewing.
Satellite growth analysis
Mycoplasmas were inoculated in four-well borosilicate glass chamber slides (Nunc Nalgene, Naperville, IL) containing SP4 medium + 3% gelatin with appropriate antibiotics. Time-lapse images were obtained at 12-h intervals per established protocols (Hasselbring et al., 2005).
Trypsin digestion
Wild-type and mutant M. pneumoniae strains cultured and harvested as described above were suspended in cold PBS buffer (pH 7.2), assayed for total protein content by bicinchoninic acid assay (Pierce, Rockford, Ill.), normalized to 1 mg/ml, and kept on ice. Aliquots of 30 µl were used per sample and time-point. As a control, some aliquots were incubated with Triton X-100 at 2% for 30 min at 37°C prior to trypsin treatment. Type XI bovine pancreas trypsin (Sigma, St. Louis, MO) and type II-0 chicken white trypsin inhibitor (Sigma, St. Louis, MO) in PBS buffer were prepared immediately prior to use. Trypsin was added to a concentration of 25 µg/ml (with PBS alone added to negative control samples) and incubated at 37°C in a water bath. At each time point, a set of tubes was removed and the reaction stopped with trypsin inhibitor (25 µg/ml; molar ratio 1.2:1 to trypsin). SDS-PAGE sample buffer was added and samples were incubated 95°C for 15 min prior to storage at −20°C. Samples were analyzed by 6% SDS-PAGE and Western immunoblotting as described previously (Hahn et al., 1996; Hasselbring et al., 2005). Rabbit polyclonal antibodies prepared against P1 protein excised from SDS-PAGE gels as described (Baseman et al., 1982), were used at 1:1,000 (anti-P1). Rabbit anti-GroEL antibodies (Proft and Herrmann, 1994) were used at 1:4,000. The secondary antibody was alkaline phosphatase-conjugated goat anti-rabbit IgG (1:7500; Promega, Madison, WI).
Statistical analysis of ultrastructural quantitations and immunoblot band intensities
Electron-dense core positioning was assessed as the ratio of the length of core extended beyond the cell body to its total length. The frequency for each ratio category was determined as a percentage of the total population. A 95% confidence interval was determined and listed for each observed frequency. Size and mean pixel intensity of TEM thin sections were determined as described (Cloward and Krause, 2010) and analyzed by ANOVA and Dunn’s Multiple Comparison post-hoc test by using GraphPad Instat, version 3.05 software (La Jolla, CA).
Digital images of immunoblots were obtained to determine protein band intensities. Each 170-kDa P1 band or pair of bands was assessed for pixel intensity using the Scion Image marquee selection tool fixed at identical dimensions for all blots, with experiments performed in triplicate. The relative pixel intensity for each band was obtained and converted to a percentage intensity of the untreated band at the same time point: ([C at t=10] / [- at t=10] × 100). Change over time of untreated samples were shown relative to t=1: ([- at t=10] / [- at t=1] × 100). No values were obtained for t=0; the values from negative controls at t=1 were used for t=0 to graphically represent the reduction between pre-treatment and t=1 for trypsin-treated samples. Differences in densitometric analysis were analyzed using multivariate ANOVA followed by Tukey HSD post-hoc pairwise comparisons of strains per time-point, by using SAS 9.2 software (The SAS Institute, Cary, NC).
Supplementary Material
Acknowledgments
We thank Mary Ard and Ed Sheppard for technical assistance and Richard Herrmann for graciously providing us with anti-GroEL antiserum. We thank Sara Satola, Amy Holst, and Myron Zaluki for their assistance with statistical analyses. This work was supported by Public Health Service research grant AI22362 from the National Institute of Allergy and Infectious Diseases to D.C.K.
References
- Balish MF, Hahn TW, Popham PL, Krause DC. Stability of Mycoplasma pneumoniae cytadherence-accessory protein HMW1 correlates with its association with the triton shell. J Bacteriol. 2001;183:3680–3688. doi: 10.1128/JB.183.12.3680-3688.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseman JB, Cole RM, Krause DC, Leith DK. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J Bacteriol. 1982;151:1514–1522. doi: 10.1128/jb.151.3.1514-1522.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biberfeld G, Biberfeld P. Ultrastructural features of Mycoplasma pneumoniae. J Bacteriol. 1970;102:855–861. doi: 10.1128/jb.102.3.855-861.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt W. Motility and multiplication of Mycoplasma pneumoniae. A phase contrast study. Pathol Microbiol (Basel) 1968;32:321–326. doi: 10.1159/000162074. [DOI] [PubMed] [Google Scholar]
- Bukau B, Reilly P, McCarty J, Walker GC. Immunogold localization of the DnaK heat shock protein in Escherichia coli cells. J Gen Microbiol. 1993;139:95–99. doi: 10.1099/00221287-139-1-95. [DOI] [PubMed] [Google Scholar]
- Cloward JM, Krause DC. Mycoplasma pneumoniae J-domain protein required for terminal organelle function. Mol Microbiol. 2009;71:1296–1307. doi: 10.1111/j.1365-2958.2009.06602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloward JM, Krause DC. Functional domain analysis of the Mycoplasma pneumoniae co-chaperone TopJ. Mol Microbiol. 2010;77:158–169. doi: 10.1111/j.1365-2958.2010.07196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallo SF, Chavoya A, Baseman JB. Characterization of the gene for a 30-kilodalton adhesion-related protein of Mycoplasma pneumoniae. Infect Immun. 1990;58:4163–4165. doi: 10.1128/iai.58.12.4163-4165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldner J, Göbel U, Bredt W. Mycoplasma pneumoniae adhesin localized to tip structure by monoclonal antibody. Nature. 1982;298:765–767. doi: 10.1038/298765a0. [DOI] [PubMed] [Google Scholar]
- Franzoso G, Hu PC, Meloni GA, Barile MF. The immunodominant 90-kilodalton protein is localized on the terminal tip structure of Mycoplasma pneumoniae. Infect Immun. 1993;61:1523–1530. doi: 10.1128/iai.61.4.1523-1530.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Gerstenecker B, Jacobs E. Topological mapping of the P1-adhesin of Mycoplasma pneumoniae with adherence-inhibiting monoclonal antibodies. J Gen Microbiol. 1990;136:471–476. doi: 10.1099/00221287-136-3-471. [DOI] [PubMed] [Google Scholar]
- Göbel U, Speth V, Bredt W. Filamentous structures in adherent Mycoplasma pneumoniae cells treated with nonionic detergents. J Cell Biol. 1981;91:537–543. doi: 10.1083/jcb.91.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn TW, Krebes KA, Krause DC. Expression in Mycoplasma pneumoniae of the recombinant gene encoding the cytadherence-associated protein HMW1 and identification of HMW4 as a product. Mol Microbiol. 1996;19:1085–1093. doi: 10.1046/j.1365-2958.1996.455985.x. [DOI] [PubMed] [Google Scholar]
- Hahn TW, Willby MJ, Krause DC. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J Bacteriol. 1998;180:1270–1276. doi: 10.1128/jb.180.5.1270-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hasselbring BM, Jordan JL, Krause DC. Mutant analysis reveals a specific requirement for protein P30 in Mycoplasma pneumoniae gliding motility. J Bacteriol. 2005;187:6281–6289. doi: 10.1128/JB.187.18.6281-6289.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselbring BM, Krause DC. Cytoskeletal protein P41 is required to anchor the terminal organelle of the wall-less prokaryote Mycoplasma pneumoniae. Mol Microbiol. 2007a;63:44–53. doi: 10.1111/j.1365-2958.2006.05507.x. [DOI] [PubMed] [Google Scholar]
- Hasselbring BM, Krause DC. Proteins P24 and P41 function in the regulation of terminal-organelle development and gliding motility in Mycoplasma pneumoniae. J Bacteriol. 2007b;189:7442–7449. doi: 10.1128/JB.00867-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedreyda CT, Krause DC. Identification of a possible cytadherence regulatory locus in Mycoplasma pneumoniae. Infect Immun. 1995;63:3479–3483. doi: 10.1128/iai.63.9.3479-3483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedreyda CT, Lee KK, Krause DC. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid. 1993;30:170–175. doi: 10.1006/plas.1993.1047. [DOI] [PubMed] [Google Scholar]
- Henderson GP, Jensen GJ. Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol Microbiol. 2006;60:376–385. doi: 10.1111/j.1365-2958.2006.05113.x. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Hartl FU. The role of molecular chaperones in protein folding. Faseb J. 1995;9:1559–1569. doi: 10.1096/fasebj.9.15.8529835. [DOI] [PubMed] [Google Scholar]
- Jordan JL, Chang HY, Balish MF, Holt LS, Bose SR, Hasselbring BM, Waldo RH, 3rd, Krunkosky TM, Krause DC. Protein P200 is dispensable for Mycoplasma pneumoniae hemadsorption but not gliding motility or colonization of differentiated bronchial epithelium. Infect Immun. 2007;75:518–522. doi: 10.1128/IAI.01344-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenri T, Seto S, Horino A, Sasaki Y, Sasaki T, Miyata M. Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J Bacteriol. 2004;186:6944–6955. doi: 10.1128/JB.186.20.6944-6955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause DC, Balish MF. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol Microbiol. 2004;51:917–924. doi: 10.1046/j.1365-2958.2003.03899.x. [DOI] [PubMed] [Google Scholar]
- Krause DC, Leith DK, Wilson RM, Baseman JB. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect Immun. 1982;35:809–817. doi: 10.1128/iai.35.3.809-817.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layh-Schmitt G, Harkenthal M. The 40- and 90-kDa membrane proteins (ORF6 gene product) of Mycoplasma pneumoniae are responsible for the tip structure formation and P1 (adhesin) association with the Triton shell. FEMS Microbiol Lett. 1999;174:143–149. doi: 10.1111/j.1574-6968.1999.tb13561.x. [DOI] [PubMed] [Google Scholar]
- Layh-Schmitt G, Herrmann R. Localization and biochemical characterization of the ORF6 gene product of the Mycoplasma pneumoniae P1 operon. Infect Immun. 1992;60:2906–2913. doi: 10.1128/iai.60.7.2906-2913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layh-Schmitt G, Herrmann R. Spatial arrangement of gene products of the P1 operon in the membrane of Mycoplasma pneumoniae. Infect Immun. 1994;62:974–979. doi: 10.1128/iai.62.3.974-979.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layh-Schmitt G, Hilbert H, Pirkl E. A spontaneous hemadsorption-negative mutant of Mycoplasma pneumoniae exhibits a truncated adhesin-related 30-kilodalton protein and lacks the cytadherence-accessory protein HMW1. J Bacteriol. 1995;177:843–846. doi: 10.1128/jb.177.3.843-846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layh-Schmitt G, Podtelejnikov A, Mann M. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiology. 2000;146(Pt 3):741–747. doi: 10.1099/00221287-146-3-741. [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman RP, Clyde WA., Jr The interrelationship of virulence, cytadsorption, and peroxide formation in Mycoplasma pneumoniae. Proc Soc Exp Biol Med. 1969;131:1163–1167. doi: 10.3181/00379727-131-34061. [DOI] [PubMed] [Google Scholar]
- Meng KE, Pfister RM. Intracellular structures of Mycoplasma pneumoniae revealed after membrane removal. J Bacteriol. 1980;144:390–399. doi: 10.1128/jb.144.1.390-399.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane D, Adan-Kubo J, Kenri T, Miyata M. Isolation and characterization of P1 adhesin, a leg protein of the gliding bacterium Mycoplasma pneumoniae. J Bacteriol. 2011;193:715–722. doi: 10.1128/JB.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft T, Herrmann R. Identification and characterization of hitherto unknown Mycoplasma pneumoniae proteins. Mol Microbiol. 1994;13:337–348. doi: 10.1111/j.1365-2958.1994.tb00427.x. [DOI] [PubMed] [Google Scholar]
- Regula JT, Boguth G, Gorg A, Hegermann J, Mayer F, Frank R, Herrmann R. Defining the mycoplasma 'cytoskeleton': the protein composition of the Triton X-100 insoluble fraction of the bacterium Mycoplasma pneumoniae determined by 2-D gel electrophoresis and mass spectrometry. Microbiology. 2001;147:1045–1057. doi: 10.1099/00221287-147-4-1045. [DOI] [PubMed] [Google Scholar]
- Romero-Arroyo CE, Jordan JL, Peacock SJ, Willby MJ, Farmer MA, Krause DC. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J Bacteriol. 1999;181:1079–1087. doi: 10.1128/jb.181.4.1079-1087.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S, Layh-Schmitt G, Kenri T, Miyata M. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J Bacteriol. 2001;183:1621–1630. doi: 10.1128/JB.183.5.1621-1630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S, Miyata M. Attachment organelle formation represented by localization of cytadherence proteins and formation of the electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J Bacteriol. 2003;185:1082–1091. doi: 10.1128/JB.185.3.1082-1091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully JG, Whitcomb RF, Clark HF, Williamson DL. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science. 1977;195:892–894. doi: 10.1126/science.841314. [DOI] [PubMed] [Google Scholar]
- Waites KB, Talkington DF. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev. 2004;17:697–728. doi: 10.1128/CMR.17.4.697-728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willby MJ, Krause DC. Characterization of a Mycoplasma pneumoniae hmw3 mutant: implications for attachment organelle assembly. J Bacteriol. 2002;184:3061–3068. doi: 10.1128/JB.184.11.3061-3068.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


