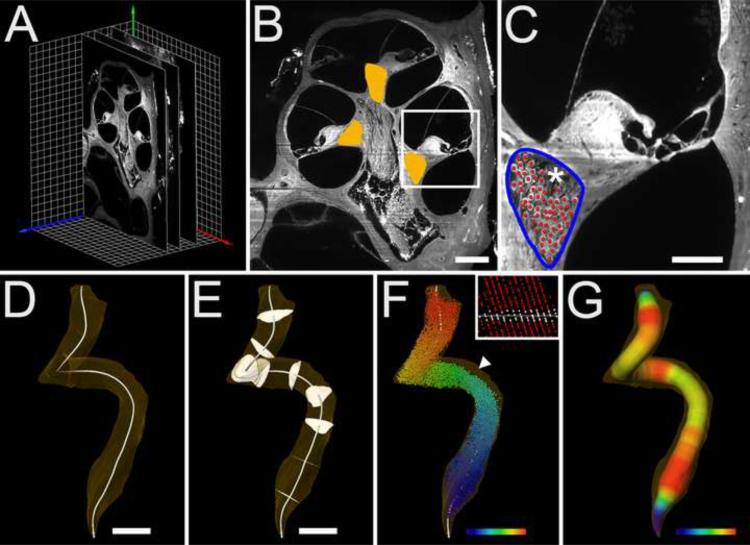
Fig. 1.
Data collection and analysis. Panel A shows three of the ~300 serial sections of a z-stack placed within a Cartesian coordinate system (red, green and blue arrows indicate x, y and z axes respectively). Each grid square is 100 × 100 μm. Panel B shows an example of Rosenthal's canal segmentation (orange overlay) in a representative mid-modiolar section. Bar indicates 200 μm. Panel C shows a lower turn from a cross section in B with the border of the canal outlined in blue and SGNs marked by manually placed red spheres using Amira. The * indicates a region of the canal that contains nerve fibers of the intraganglionic spiral bundle. It was this type of cross section that was used to determine the reproducibility of RC area by two observers. Bar indicates 100 μm. Panel D shows a 3D rendering of Rosenthal's canal (transparent orange) with B-spline generated centroid (white line). Bar indicates 200 μm. Panel E shows the orientation of orthogonal sections distributed every 10% distance from base to apex (10–90%). Bar indicates 200 μm. Panel F shows spheres representing SGN cell bodies color mapped by base-apex location along the centroid. Color gradient indicates the base (purple), middle (green, yellow-green) and apex (red). The clear area of RC (arrowhead) indicates the position of the intraganglionic spiral bundle within the canal. SGNs were counted in serial sections as shown in C. Panel F (insert) indicates the plane of an orthogonal section which in this case bisects 14 original imaging planes through the canal. Orthogonal plane at this percent distance location is represented by a white line. Panel G shows a representation of the canal's radius measured as the Euclidian distance from the exterior of the canal to its centroid. A color gradient map indicates radii between 20 μm (purple) and 65 μm (red).