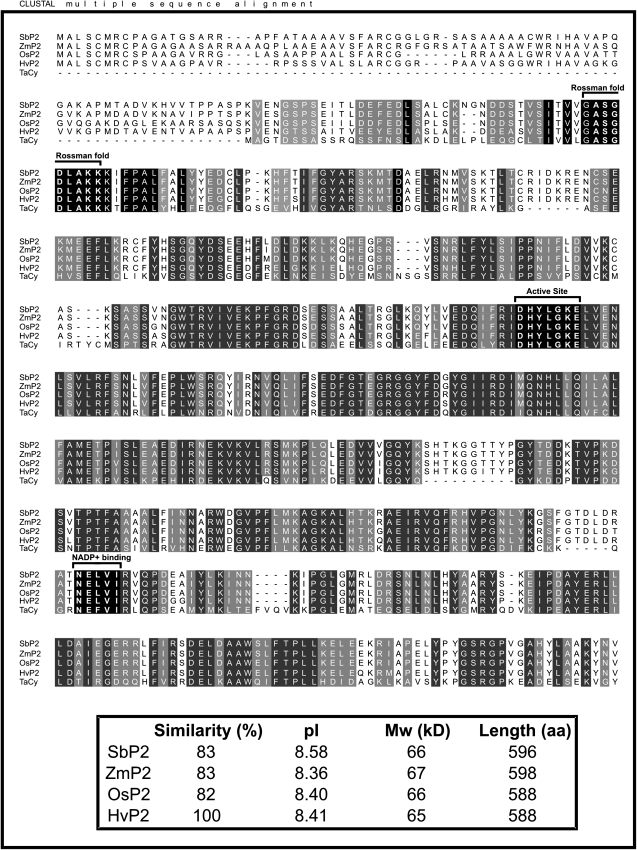
Fig. 2.
Amino acid sequence alignment of P2-G6PDHs from Hordeum vulgare (Hv, CAL44728), Oryza sativa (Os, LOC_Os07g22350), Zea mays (Zm, ACG29334), Sorghum bicolor (Sb, estExt_Genewise1Plus.C_chr_60876), and Triticum aestivum (Ta, BAA97663). The alignment was performed with ClustalW. Strictly conserved amino acids are on a black background, while functionally conserved amino acids are on a grey background. The active site (DHYLGKEL), the Rossman-fold motif (ASGDLAKK), and the core of the NADP+-binding site (NELVI) are indicated by brackets. In the lower box, the percentage similarity, deduced isoelectric points (pI), molecular weights (MW), and protein lengths in amino acids (aa) of the different sequences are shown.