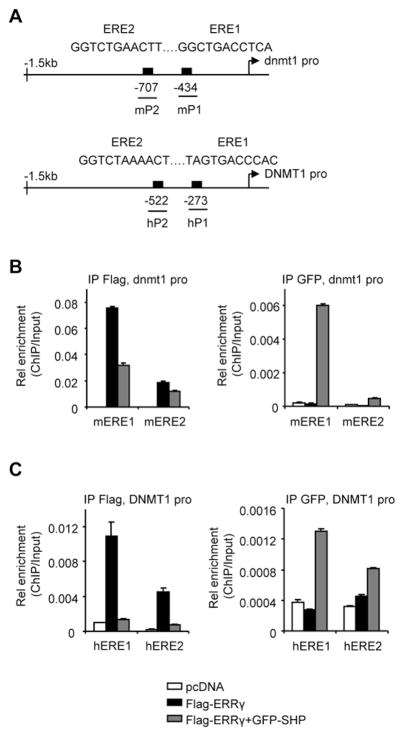
Fig. 2.
SHP inhibition of ERRγ recruitment to the Dnmt1 promoter. (A) Schematic showing the putative ERRγ binding sites (EREs) in the dnmt1 and DNMT1 promoters. (B–C) ChIP assays to determine ERRγ and SHP co-immunoprecipitation (Co-IP) to the ERE region of the dnmt1 (B) or DNMT1 (C) promoter. ERRγ and SHP were exogenously expressed in Hela cells with Flag-ERRγ (5 μg, 10 cm plate) and/or GFP-SHP (5 μg, 10 cm plate) plasmids, along with the dnmt1 promoter. The chromatin was co-immunoprecipitated using anti-Flag or anti-GFP antibodies. Real-time qPCR was used to determine the association of ERRγ and SHP to the exogenous dnmt1 promoter using mP1 and mP2 primers (B), and their association to the endogenous DNMT1 promoter was determined by hP1 and hP2 primers (C). PCR products were amplified from input-positive controls, IgG-negative controls, and antibody immunoprecipitates. Histograms show antibody/input ratios of PCR products quantified using qPCR, expressed as Relative (Rel) enrichment. Error bars represent SEM from three independent measurements.