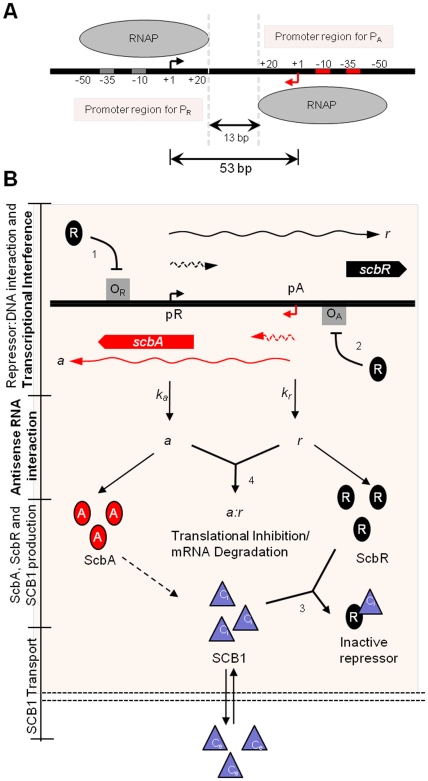
Figure 1. Convergent transcription in the scbA-scbR gene regulatory network.
(A) Schematic of the promoter regions for pA and pR, and RNAP footprint at the respective promoters. (B) The scbA-scbR gene regulatory network. Convergent promoter pA-pR drive expression of genes scbA (shown in red) and scbR (shown in black) present on the opposite DNA strands (shown by black lines) to give rise to full-length transcripts a and r (RNA denoted by curved lines) and short truncated RNA (denoted by dashed-curved lines) respectively. Transcriptional Interference model: Collision between elongating RNAPs and between an elongating RNAP and a stationary RNAP at the opposing promoter causes transcriptional termination and results in the generation of short truncated transcripts (dashed-curved lines) from promoters pA and pR respectively. Full-length transcripts a and r result when elongating RNAPs escape collision. Antisense Regulation: Hybrid RNA complexes formed between full-length a and r RNA result in translational inhibition or mRNA degradation. Protein ScbR (R) can repress transcription from pA and pR (indicated by blunt arrows) by binding to operator sites OA and OR adjacent to promoters pA and pR respectively. Only full-length transcripts a and r are translated to protein ScbA (A) and ScbR (R). The intracellular γ-butyrolactone SCB1 (denoted by Ci) is produced from glycerol derivatives and β-keto acid by the enzymatic action of γ-butyrolactone synthase ScbA. SCB1 forms complex with ScbR (CiR) to sequester its repressive effect. SCB1 diffuses out of the cell in to the extracellular environment and vice versa (denoted by Ce). The reactions are numbered according to equations in Table 1.