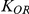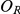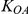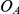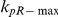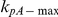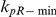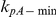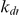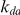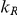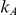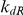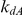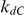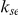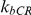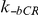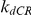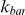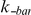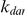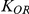
|
Equilibrium binding constant of ScbR to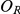
|
8.82 |
0.44 |
37.92 |
[69], [70]
|
nM |
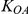
|
Equilibrium Binding constant of ScbR to 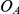
|
2.68 |
0.12 |
4.92 |
[69], [70]
|
nM |
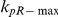
|
Transcription from PR in de-repressed state |
1.5×10−1
|
7.5×10−3
|
3.0×10−1
|
[65]
|
s−1
|
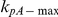
|
Transcription from PA in de-repressed state |
4.5×10−1
|
2.25×10−2
|
9.0×10−1
|
[65]
|
s−1
|
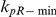
|
Transcription from PR in repressed state |
1.0×10−3
|
5.0×10−5
|
3.0×10−3
|
[65]
|
s−1
|
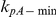
|
Transcription from PA in repressed state |
8.0×10−4
|
4.0×10−5
|
2.8×10−3
|
[65]
|
s−1
|
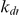
|
Degradation of full-length r RNA |
7.0×10−3
|
3.5×10−4
|
1.4×10−2
|
[72]
|
s−1
|
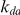
|
Degradation of full-length a RNA |
8.1×10−4
|
4.1×10−4
|
1.6×10−2
|
[72]
|
s−1
|
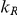
|
ScbR protein translation |
3.6×10−1
|
9.0×10−2
|
3.8×10−1
|
[68], [69]
|
s−1
|
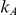
|
ScbA protein translation |
6.6×10−2
|
1.0×10−2
|
1.2×10−1
|
[68], [69]
|
s−1
|
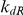
|
ScbR protein degradation |
4.0×10−3
|
1.0×10−3
|
8.0×10−3
|
[69]
|
s−1
|
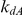
|
ScbA protein degradation |
1.8×10−3
|
8.0×10−4
|
2.0×10−3
|
[69]
|
s−1
|
|
µ
|
growth rate |
6.0×10−5
|
6.0×10−5
|
8.0×10−5
|
[71]
|
s−1
|
|
kC
|
SCB1 synthesis |
7.4×10−1
|
7.4×10−2
|
37 |
[66]
|
s−1
|
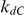
|
SCB1 degradation |
6.7×10−5
|
6.7×10−6
|
6.7×10−3
|
Max. half life 1 hr. |
s−1
|
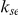
|
SCB1 secretion |
8.3×10−2
|
8.3×10−2
|
4.2 |
[67]
|
s−1
|
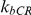
|
Binding of ScbR and SCB1 to form SCB1-ScbR complex |
8.3×10−2
|
4.2×10−3
|
2.53×10−1
|
[70]
|
nM−1 s−1
|
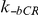
|
Unbinding of SCB1-ScbR complex |
1.7×102
|
8.5 |
1.95×102
|
[70]
|
s−1
|
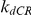
|
SCB1-ScbR degradation |
3.1×10−3
|
3.1×10−3
|
6.8×10−2
|
[70]
|
s−1
|
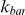
|
Binding rate constant of full-length (a, r) to form hybrid RNA complexes a:r
|
1.0×10−3
|
6.5×10−4
|
1.6×10−1
|
[48]
|
nM−1s−1
|
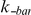
|
Unbinding of ar complex |
1.0×10−2
|
0 |
2.0×10−1
|
[48]
|
s−1
|
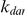
|
RNA hybrid complex ar degradation |
1×10−2
|
0 |
1.24 |
[48]
|
s−1
|