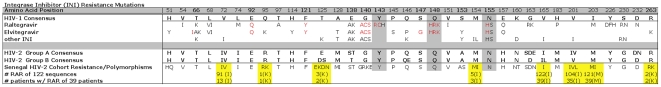
Figure 2. HIV-2 integrase variation at sites implicated in INI resistance.
(Top) Amino acid substitutions that appear in HIV-1 sequences from patients treated with raltegravir, elvitegravir, or other INIs, or that emerge in HIV-1 in response to INI selection in culture. This list was compiled from sources listed under Methods. The consensus HIV-1 sequence (http://www.hiv.lanl.gov) is shown for comparison. Gray boxes indicate the locations of primary INI resistance mutations in HIV-1. Amino acid changes that are associated with >5-fold resistance to raltegravir or elvitegravir in HIV-1 (http://hivdb.stanford.edu) are shown in red. (Bottom) Consensus sequences for group A and group B HIV-2 isolates were derived from previously-published data (http://www.hiv.lanl.gov); positions that contain two or more residues are polymorphic in HIV-2. Patient-derived HIV-2 integrase sequences (n = 122) were obtained from 39 INI-naïve individuals in the Senegal cohort. HIV-2 integrase residues that correspond to INI resistance-associated mutations in HIV-1 are highlighted in yellow boxes. Values below the boxes indicate the number of HIV-2 patient sequences containing the each resistance-associated residue (RAR, in parentheses) and the number of patients in which these amino acids were present.