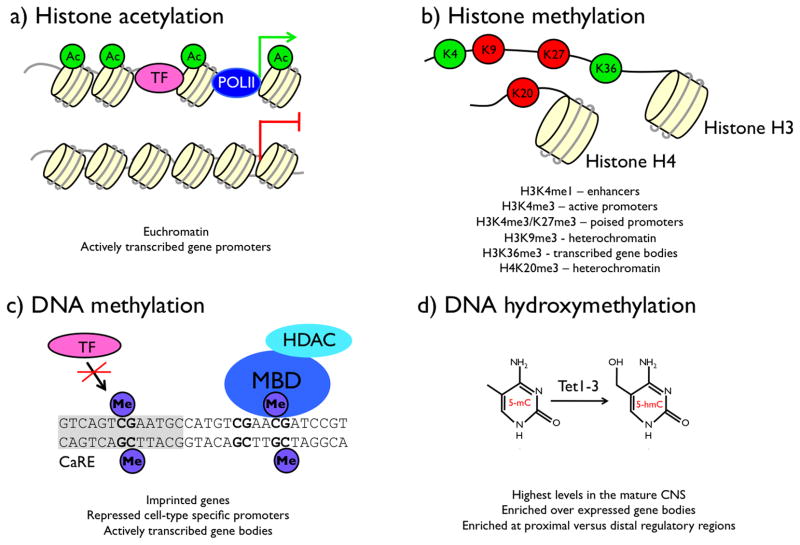
FIGURE 5. Chromatin modifications that regulate transcription.
Post-translational modifications of histones and covalent modifications of genomic DNA regulate transcription. a) Acetylation (Ac) can be added to multiple lysine residues in the N-terminal tail domain of histones H3 and H4. This modification is preferentially associated with transcriptionally active genes. b) Methylation (me) of histones H3 and H4 occurs at specific lysine (K) and arginine residues. The functional consequence of each of these modifications depends on the specific amino acid modified (K4, K9, K27, and K36 in histone H3, K20 in histone H4) (Kouzarides, 2002). Sites of methylation associated with transcriptional activation are shown in green, sites associated with transcriptional repression are shown in red. Each lysine can be mono- (me1), di- (me2), or trimethylated (me3), and the global distribution of these methyl marks varies across different kinds of genetic regulatory elements (Hon et al., 2009; Mikkelsen et al., 2007). c) In differentiated mammalian cells, DNA methylation occurs on cytosines at a subset of CpG dinucleotides. Methylation can regulate transcription by sterically blocking the association of a transcription factor (TF) with its binding site (the gray box represents a CaRE), or by recruiting the association of a protein with a methyl-DNA binding domain (MBD), which can act as a scaffold for additional chromatin regulatory enzymes. d) 5-hydroxymethylation of cytosine (5-hmC) is catalyzed by the enzymes Tet1, Tet2, and Tet3, which add a hydroxyl group to methylated cytosine bases (5-mC) (Ito et al., 2010; Ko et al., 2010; Tahiliani et al., 2009). Little is known about the functional relevance of this modification of DNA, but new chemical methods are beginning to allow its genome-wide distribution to be described (Song et al., 2011).