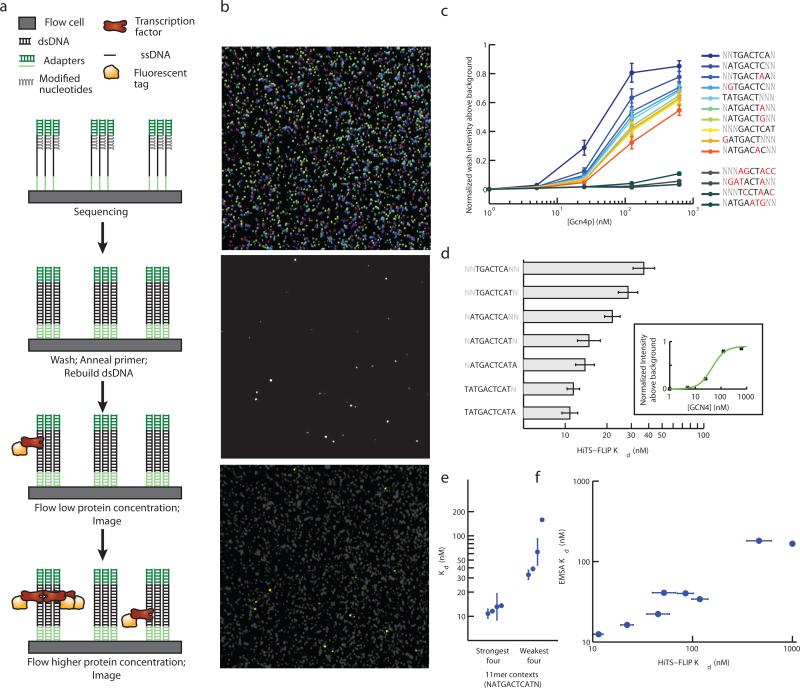
Figure 1. High-throughput sequencing fluorescent ligand interaction profile (HiTS-FLIP) method.
a) HiTS-FLIP method schematic. A microfluidic flow cell with anchored single-stranded DNA is sequenced by synthesis. Second-strand DNA is stripped and rebuilt using Klenow and unmodified dNTPs to form dsDNA clusters. Fluorescently labeled Gcn4p is introduced at different concentrations and binding is imaged. b) Partial images from sequencing cycles (false colored for each nucleotide, top), a Gcn4p binding cycle (center), and a merge of the top and center with Gcn4p clusters highlighted in yellow (lower). The images shown represent roughly 0.003% of a flow cell. c) Intensity of binding versus Gcn4p concentration for the top ten 7mers (see text) and four 7mers with expected binding affinity near zero. Mismatches from the consensus sequence are marked in red. Error bars indicate 95% confidence intervals based on estimates from 5 flow cell lanes. d) Dissociation constants (Kd values) calculated by fitting a Hill equation to intensity measurements (inset; for TGACTCA). Top 7mer by Kd as well as selected extensions that had significantly increased affinity are shown. Error bars indicate standard deviation estimated from comparison of 5 flow cell lanes. e) Kd (mean and standard error) for 11mers containing the strongest-binding 9mer sequence, ATGACTCAT. The four 11mers with the strongest Kd (flanked by T,A; T,C; G,A; and C,A) and the four with the weakest Kd (flanked by C,T; A,G; A,C; and C,G) are shown. f) Dissociation constants measured by HiTS-FLIP (mean and standard error of 5 flow cell lanes) compared with those measured by EMSA (mean of duplicate experiments). Nine sequences chosen to have a wide range of expected binding affinity were assayed.