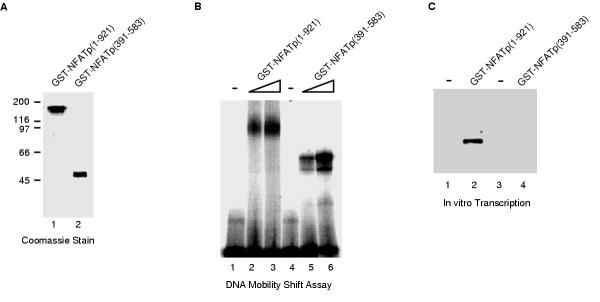
Figure 4.
Transcriptional activation by NFATp in a reconstituted transcription system is dependent on regions outside of the DNA binding domain. (A) GST-fusions to full-length NFATp (amino acids 1-921) and the DNA-binding domain of NFATp (amino acids 391-583) were expressed in insect cells, purified by affinity chromatography, and eluted with glutathione. The purified proteins were analyzed by SDS-PAGE and stained with coomassie brilliant blue. (B) GST-NFATp(1-921) and GST-NFATp(391-583) are active for binding DNA. Proteins were incubated with a double stranded oligonucleotide containing a consensus NFAT-binding site. Protein/DNA complexes and free DNA were resolved on a native 4% polyacrylamide gel, and detected by autoradiography. The final concentrations of GST-NFATp(1-921) and GST-NFATp(391-583) were: 17 nM (lanes 2 and 5) and 35 nM (lanes 3 and 6). (C) GST-NFATp(1-921), but not GST-NFATp(391-583) activates transcription in a reconstituted transcription system. Transcription reactions were performed in a reconstituted RNA polymerase II transcription system using a DNA template consisting of three high affinity NFAT sites upstream of the adenovirus major late core promoter and a 200 bp G-less cassette. 40 ng of either GST-NFATp(1-921) and GST-NFATp(391-583) were added to reactions as indicated. Transcripts were purified, resolved by denaturing PAGE, and visualized with a PhosphorImager.