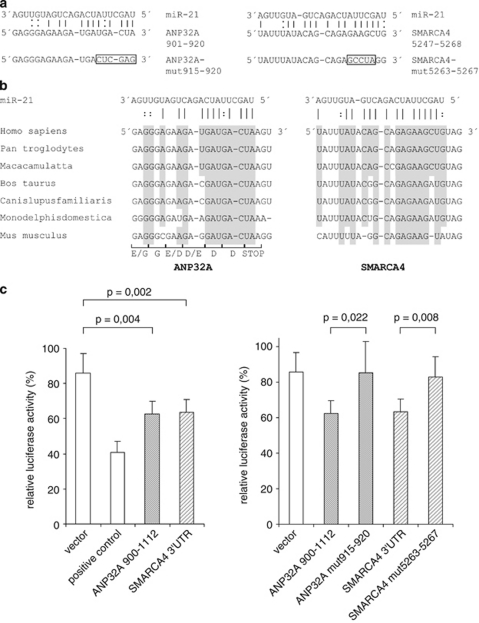
Figure 4.
ANP32A and SMARCA4 are direct targets of miR-21. (a) Putative miR-21-binding sites within ANP32A (NM_006305) and SMARCA4 (NM_001128849) genes. Perfect matches are indicated by a line; G:U pairs by a colon. The frame marks nucleotides mutated for the reporter gene assays. (b) Comparison of nucleotides between the miR-21 sequence and its targets in various species. For the ANP32A site overlapping the open reading frame, amino acids are indicated. (c) Cloned ANP32A and SMARCA4sequences are targeted by miR-21 (left panel), and mutation of the putative binding sites abolishes this effect to wild type level (right panel). pMIR-REPORT vectors containing ANP32A-900-1112, SMARCA4-3′ untranslated region (3′UTR) and their mutated variants were co-transfected with miR-21 or control precursor into LNCaP cells. A vector containing three miR-21-binding sites was used as a positive control. Luciferase activity was assayed after 48 h. Data are normalized to the corresponding control sample. Values are the means±s.d. from six independent experiments. The P-values were determined using a two-tailed t-test.