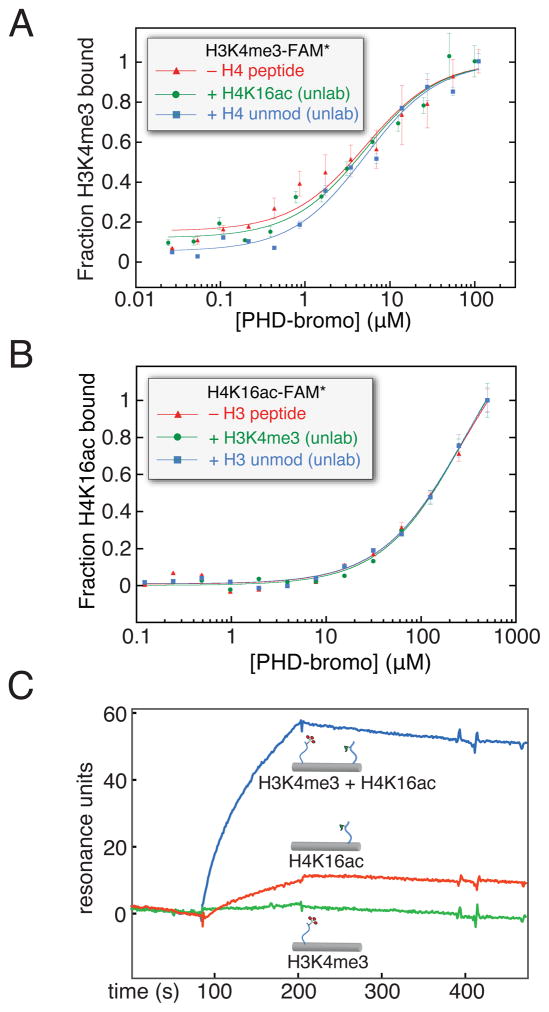
Figure 2. Is there allostery or bivalent cooperativity in simultaneous ligand binding by the BPTF PHD-bromodomain?
(A) The possible allosteric cooperative binding at the peptide level is examined by fluorescence polarization anisotropy using 100 nM fluorescein-labelled H3K4me3 peptide, with either no additional peptide, excess unlabeled H4K16ac peptide (500 μM), or excess unlabeled H4 peptide without acetylation (500 μM). (B) The converse experiment with respect to panel A: with unlabeled H3K4me3 in excess (20 μM, ~10-fold above PHD-H3K4me3 Kd) and fluorescein-H4K16ac (150 nM), protein is titrated and resulting fluorescence polarization anisotropy measured (expressed here as fraction bound). (C) A dsDNA scaffold, selected for rigidity while retaining little predicted bending, was used to covalently install H3K4me3 (H3K4me3[1–8], green), H4K16ac (H4K16ac [12–20], red), or both peptides (blue) at specific positions by disulfide formation with cystamine derivatized convertible dC nucleosides (Figure S2). All of these DNA-protein conjugates were immobilized in different flow channels via a single 3′-biotin linkage to a streptavidin coated surface plasmon resonance chip at low density, untagged PHD-bromo was applied and background binding was subtracted from an empty flow cell. See also Figure S2. (data are represented as mean ± SD)